Feature selection for high dimensional microarray gene expression data via weighted signal to noise ratio
- PMID: 37098036
- PMCID: PMC10128961
- DOI: 10.1371/journal.pone.0284619
Feature selection for high dimensional microarray gene expression data via weighted signal to noise ratio
Abstract
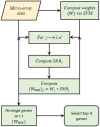
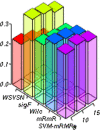
Feature selection in high dimensional gene expression datasets not only reduces the dimension of the data, but also the execution time and computational cost of the underlying classifier. The current study introduces a novel feature selection method called weighted signal to noise ratio (WSNR) by exploiting the weights of features based on support vectors and signal to noise ratio, with an objective to identify the most informative genes in high dimensional classification problems. The combination of two state-of-the-art procedures enables the extration of the most informative genes. The corresponding weights of these procedures are then multiplied and arranged in decreasing order. Larger weight of a feature indicates its discriminatory power in classifying the tissue samples to their true classes. The current method is validated on eight gene expression datasets. Moreover, results of the proposed method (WSNR) are also compared with four well known feature selection methods. We found that the (WSNR) outperform the other competing methods on 6 out of 8 datasets. Box-plots and Bar-plots of the results of the proposed method and all the other methods are also constructed. The proposed method is further assessed on simulated data. Simulation analysis reveal that (WSNR) outperforms all the other methods included in the study.
Copyright: © 2023 Hamraz et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

















References
MeSH terms
LinkOut - more resources
Full Text Sources

