The Combined Effect of Hg(II) Speciation, Thiol Metabolism, and Cell Physiology on Methylmercury Formation by Geobacter sulfurreducens
- PMID: 37098211
- PMCID: PMC10173453
- DOI: 10.1021/acs.est.3c00226
The Combined Effect of Hg(II) Speciation, Thiol Metabolism, and Cell Physiology on Methylmercury Formation by Geobacter sulfurreducens
Abstract
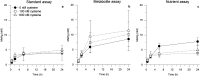
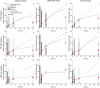
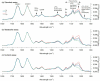
The chemical and biological factors controlling microbial formation of methylmercury (MeHg) are widely studied separately, but the combined effects of these factors are largely unknown. We examined how the chemical speciation of divalent, inorganic mercury (Hg(II)), as controlled by low-molecular-mass thiols, and cell physiology govern MeHg formation by Geobacter sulfurreducens. We compared MeHg formation with and without addition of exogenous cysteine (Cys) to experimental assays with varying nutrient and bacterial metabolite concentrations. Cysteine additions initially (0-2 h) enhanced MeHg formation by two mechanisms: (i) altering the Hg(II) partitioning from the cellular to the dissolved phase and/or (ii) shifting the chemical speciation of dissolved Hg(II) in favor of the Hg(Cys)2 complex. Nutrient additions increased MeHg formation by enhancing cell metabolism. These two effects were, however, not additive since cysteine was largely metabolized to penicillamine (PEN) over time at a rate that increased with nutrient addition. These processes shifted the speciation of dissolved Hg(II) from complexes with relatively high availability, Hg(Cys)2, to complexes with lower availability, Hg(PEN)2, for methylation. This thiol conversion by the cells thereby contributed to stalled MeHg formation after 2-6 h Hg(II) exposure. Overall, our results showed a complex influence of thiol metabolism on microbial MeHg formation and suggest that the conversion of cysteine to penicillamine may partly suppress MeHg formation in cysteine-rich environments like natural biofilms.
Keywords: anaerobe microorganisms; low molecular mass thiols; mercury methylation; mercury speciation.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Microbial Biosynthesis of Thiol Compounds: Implications for Speciation, Cellular Uptake, and Methylation of Hg(II).Environ Sci Technol. 2019 Jul 16;53(14):8187-8196. doi: 10.1021/acs.est.9b01502. Epub 2019 Jul 1. Environ Sci Technol. 2019. PMID: 31257868
-
Effects of Cellular Sorption on Mercury Bioavailability and Methylmercury Production by Desulfovibrio desulfuricans ND132.Environ Sci Technol. 2016 Dec 20;50(24):13335-13341. doi: 10.1021/acs.est.6b04041. Epub 2016 Nov 29. Environ Sci Technol. 2016. PMID: 27993064
-
Mercury methylation by Geobacter metallireducens GS-15 in the presence of Skeletonema costatum.Sci Total Environ. 2019 Jun 25;671:208-214. doi: 10.1016/j.scitotenv.2019.03.222. Epub 2019 Mar 15. Sci Total Environ. 2019. PMID: 30928750
-
Microbial Mercury Methylation in Aquatic Environments: A Critical Review of Published Field and Laboratory Studies.Environ Sci Technol. 2019 Jan 2;53(1):4-19. doi: 10.1021/acs.est.8b02709. Epub 2018 Dec 21. Environ Sci Technol. 2019. PMID: 30525497 Review.
-
Mercury methylation in rice paddy and accumulation in rice plant: A review.Ecotoxicol Environ Saf. 2020 Jun 1;195:110462. doi: 10.1016/j.ecoenv.2020.110462. Epub 2020 Mar 13. Ecotoxicol Environ Saf. 2020. PMID: 32179234 Review.
Cited by
-
Anaerobic HgII reduction is driven by cellular HgII-thiol interactions.Access Microbiol. 2025 Jan 28;7(1):000932.v3. doi: 10.1099/acmi.0.000932.v3. eCollection 2025. Access Microbiol. 2025. PMID: 40698175 Free PMC article.
-
Biochemical characterization and mercury methylation capacity of Geobacter sulfurreducens biofilms grown in media containing iron hydroxide or fumarate.Biofilm. 2023 Jul 29;6:100144. doi: 10.1016/j.bioflm.2023.100144. eCollection 2023 Dec 15. Biofilm. 2023. PMID: 37583615 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

