Diverse Molecular Mechanisms Underlying Microbe-Inducing Male Killing in the Moth Homona magnanima
- PMID: 37098937
- PMCID: PMC10231181
- DOI: 10.1128/aem.02095-22
Diverse Molecular Mechanisms Underlying Microbe-Inducing Male Killing in the Moth Homona magnanima
Abstract
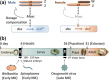
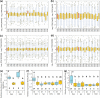
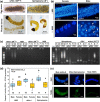
Male killing (MK) is a type of reproductive manipulation induced by microbes, where sons of infected mothers are killed during development. MK is a strategy that enhances the fitness of the microbes, and the underlying mechanisms and the process of their evolution have attracted substantial attention. Homona magnanima, a moth, harbors two embryonic MK bacteria, namely, Wolbachia (Alphaproteobacteria) and Spiroplasma (Mollicutes), and a larval MK virus, Osugoroshi virus (OGV; Partitiviridae). However, whether the three distantly related male killers employ similar or different mechanisms to accomplish MK remains unknown. Here, we clarified the differential effects of the three male killers on the sex-determination cascades and development of H. magnanima males. Reverse transcription-PCR demonstrated that Wolbachia and Spiroplasma, but not OGVs, disrupted the sex-determination cascade of males by inducing female-type splice variants of doublesex (dsx), a downstream regulator of the sex-determining gene cascade. We also found that MK microbes altered host transcriptomes in different manners; Wolbachia impaired the host dosage compensation system, whereas Spiroplasma and OGVs did not. Moreover, Wolbachia and Spiroplasma, but not OGVs, triggered abnormal apoptosis in male embryos. These findings suggest that distantly related microbes employ distinct machineries to kill males of the identical host species, which would be the outcome of the convergent evolution. IMPORTANCE Many microbes induce male killing (MK) in various insect species. However, it is not well understood whether microbes adopt similar or different MK mechanisms. This gap in our knowledge is partly because different insect models have been examined for each MK microbe. Here, we compared three taxonomically distinct male killers (i.e., Wolbachia, Spiroplasma, and a partiti-like virus) that infect the same host. We provided evidence that microbes can cause MK through distinct mechanisms that differ in the expression of genes involved in sex determination, dosage compensation, and apoptosis. These results imply independent evolutionary scenarios for the acquisition of their MK ability.
Keywords: Partitiviridae; Spiroplasma; Wolbachia; endosymbionts; male killing; symbiosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Prophage-encoded Hm-oscar gene recapitulates Wolbachia-induced male-killing in the tea tortrix moth Homona magnanima.Elife. 2025 Apr 14;13:RP101101. doi: 10.7554/eLife.101101. Elife. 2025. PMID: 40227227 Free PMC article.
-
Identification of an Early Male-Killing Agent in the Oriental Tea Tortrix, Homona magnanima.J Hered. 2017 Jul 1;108(5):553-560. doi: 10.1093/jhered/esx049. J Hered. 2017. PMID: 28505369
-
Coexistence of Two Male-Killers and Their Impact on the Development of Oriental Tea Tortrix Homona magnanima.Microb Ecol. 2021 Jan;81(1):193-202. doi: 10.1007/s00248-020-01566-x. Epub 2020 Aug 1. Microb Ecol. 2021. PMID: 32737539
-
Molecular Biology of Cytoplasmic Incompatibility Caused by Wolbachia Endosymbionts.Annu Rev Microbiol. 2023 Sep 15;77:299-316. doi: 10.1146/annurev-micro-041020-024616. Epub 2023 Jun 7. Annu Rev Microbiol. 2023. PMID: 37285552 Review.
-
The Spiroplasma heritable bacterial endosymbiont of Drosophila.Fly (Austin). 2010 Jan-Mar;4(1):80-7. doi: 10.4161/fly.4.1.10883. Epub 2010 Jan 5. Fly (Austin). 2010. PMID: 20081357 Review.
Cited by
-
Infection pattern of male-killing viruses alters phenotypes in the tea tortrix moth Homona magnanima.Heredity (Edinb). 2025 Feb;134(2):120-128. doi: 10.1038/s41437-024-00741-x. Epub 2024 Dec 26. Heredity (Edinb). 2025. PMID: 39725691
-
Endosymbionts interacting with sex-determining genes and processes.Curr Opin Insect Sci. 2025 Jul 12;72:101410. doi: 10.1016/j.cois.2025.101410. Online ahead of print. Curr Opin Insect Sci. 2025. PMID: 40659090 Review.
-
Cell-based assays and comparative genomics revealed the conserved and hidden effects of Wolbachia on insect sex determination.PNAS Nexus. 2024 Aug 22;3(9):pgae348. doi: 10.1093/pnasnexus/pgae348. eCollection 2024 Sep. PNAS Nexus. 2024. PMID: 39228812 Free PMC article.
-
Two male-killing Wolbachia from Drosophila birauraia that are closely related but distinct in genome structure.R Soc Open Sci. 2024 Jan 10;11(1):231502. doi: 10.1098/rsos.231502. eCollection 2024 Jan. R Soc Open Sci. 2024. PMID: 38204789 Free PMC article.
-
Prophage-encoded Hm-oscar gene recapitulates Wolbachia-induced male-killing in the tea tortrix moth Homona magnanima.Elife. 2025 Apr 14;13:RP101101. doi: 10.7554/eLife.101101. Elife. 2025. PMID: 40227227 Free PMC article.
References
-
- Hurst GDD, Majerus ME. 1993. Why do maternally inherited microorganisms kill males? Heredity 71:81–95. doi:10.1038/hdy.1993.110. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

