PlyKp104, a Novel Phage Lysin for the Treatment of Klebsiella pneumoniae, Pseudomonas aeruginosa, and Other Gram-Negative ESKAPE Pathogens
- PMID: 37098944
- PMCID: PMC10190635
- DOI: 10.1128/aac.01519-22
PlyKp104, a Novel Phage Lysin for the Treatment of Klebsiella pneumoniae, Pseudomonas aeruginosa, and Other Gram-Negative ESKAPE Pathogens
Abstract
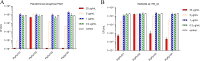
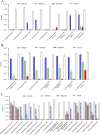
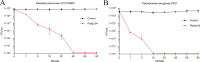
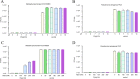
Klebsiella pneumoniae and Pseudomonas aeruginosa are two leading causes of burn and wound infections, pneumonia, urinary tract infections, and more severe invasive diseases, which are often multidrug resistant (MDR) or extensively drug resistant. Due to this, it is critical to discover alternative antimicrobials, such as bacteriophage lysins, against these pathogens. Unfortunately, most lysins that target Gram-negative bacteria require additional modifications or outer membrane permeabilizing agents to be bactericidal. We identified four putative lysins through bioinformatic analysis of Pseudomonas and Klebsiella phage genomes in the NCBI database and then expressed and tested their intrinsic lytic activity in vitro. The most active lysin, PlyKp104, exhibited >5-log killing against K. pneumoniae, P. aeruginosa, and other Gram-negative representatives of the multidrug-resistant ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, K. pneumonia, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species) without further modification. PlyKp104 displayed rapid killing and high activity over a wide pH range and in high concentrations of salt and urea. Additionally, pulmonary surfactants and low concentrations of human serum did not inhibit PlyKp104 activity in vitro. PlyKp104 also significantly reduced drug-resistant K. pneumoniae >2 logs in a murine skin infection model after one treatment of the wound, suggesting that this lysin could be used as a topical antimicrobial against K. pneumoniae and other MDR Gram-negative infections.
Keywords: ESKAPE; Gram negative; Klebsiella; Pseudomonas; antibiotic resistance; bacteriophage; endolysin; lysin; skin infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

