The Pseudomonas aeruginosa Biofilm Matrix Protein CdrA Has Similarities to Other Fibrillar Adhesin Proteins
- PMID: 37098957
- PMCID: PMC10210978
- DOI: 10.1128/jb.00019-23
The Pseudomonas aeruginosa Biofilm Matrix Protein CdrA Has Similarities to Other Fibrillar Adhesin Proteins
Abstract
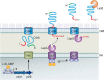
The ability of bacteria to adhere to each other and both biotic and abiotic surfaces is key to biofilm formation, and one way that bacteria adhere is using fibrillar adhesins. Fibrillar adhesins share several key characteristics, including (i) they are extracellular, surface-associated proteins, (ii) they contain an adhesive domain as well as a repetitive stalk domain, and (iii) they are either a monomer or homotrimer (i.e., identical, coiled-coil) of a high molecular weight protein. Pseudomonas aeruginosa uses the fibrillar adhesin called CdrA to promote bacterial aggregation and biofilm formation. Here, the current literature on CdrA is reviewed, including its transcriptional and posttranslational regulation by the second messenger c-di-GMP as well as what is known about its structure and ability to interact with other molecules. I highlight its similarities to other fibrillar adhesins and discuss open questions that remain to be answered toward a better understanding of CdrA.
Keywords: CdrA; Pseudomonas aeruginosa; biofilms; fibrillar adhesin.
Conflict of interest statement
The author declares no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

