Splicing Factor SRSF1 Promotes Pancreatitis and KRASG12D-Mediated Pancreatic Cancer
- PMID: 37098965
- PMCID: PMC10330071
- DOI: 10.1158/2159-8290.CD-22-1013
Splicing Factor SRSF1 Promotes Pancreatitis and KRASG12D-Mediated Pancreatic Cancer
Abstract
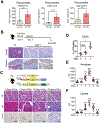
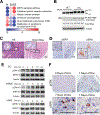
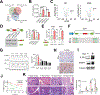
Inflammation is strongly associated with pancreatic ductal adenocarcinoma (PDAC), a highly lethal malignancy. Dysregulated RNA splicing factors have been widely reported in tumorigenesis, but their involvement in pancreatitis and PDAC is not well understood. Here, we report that the splicing factor SRSF1 is highly expressed in pancreatitis, PDAC precursor lesions, and tumors. Increased SRSF1 is sufficient to induce pancreatitis and accelerate KRASG12D-mediated PDAC. Mechanistically, SRSF1 activates MAPK signaling-partly by upregulating interleukin 1 receptor type 1 (IL1R1) through alternative-splicing-regulated mRNA stability. Additionally, SRSF1 protein is destabilized through a negative feedback mechanism in phenotypically normal epithelial cells expressing KRASG12D in mouse pancreas and in pancreas organoids acutely expressing KRASG12D, buffering MAPK signaling and maintaining pancreas cell homeostasis. This negative feedback regulation of SRSF1 is overcome by hyperactive MYC, facilitating PDAC tumorigenesis. Our findings implicate SRSF1 in the etiology of pancreatitis and PDAC, and point to SRSF1-misregulated alternative splicing as a potential therapeutic target.
Significance: We describe the regulation of splicing factor SRSF1 expression in the context of pancreas cell identity, plasticity, and inflammation. SRSF1 protein downregulation is involved in a negative feedback cellular response to KRASG12D expression, contributing to pancreas cell homeostasis. Conversely, upregulated SRSF1 promotes pancreatitis and accelerates KRASG12D-mediated tumorigenesis through enhanced IL1 and MAPK signaling. This article is highlighted in the In This Issue feature, p. 1501.
©2023 American Association for Cancer Research.
Conflict of interest statement
ARK is a co-founder, Director, Chair of the SAB, and shareholder of Stoke Therapeutics. ARK is on the SABs and holds shares of Skyhawk Therapeutics, Envisagenics, Assembl.cns, and Autoimmunity Biologic Solutions, and is a consultant for Biogen. DAT is on the SAB and holds shares with Leap Therapeutics, Surface Oncology, Sonata, and Mestag Therapeutics. DAT is a scientific co-founder of Mestag Therapeutics. DAT receives research support from Mestag Therapeutics and ONO therapeutics.
Figures



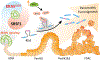
References
-
- Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. The Lancet 2020;395:2008–20. - PubMed
-
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003;4:437–50. - PubMed
-
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7:469–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

