e-RNA: a collection of web-servers for the prediction and visualisation of RNA secondary structure and their functional features
- PMID: 37099369
- PMCID: PMC10320080
- DOI: 10.1093/nar/gkad296
e-RNA: a collection of web-servers for the prediction and visualisation of RNA secondary structure and their functional features
Abstract
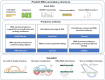
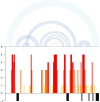
e-RNA is a collection of web-servers for the prediction and visualisation of RNA secondary structures and their functional features, including in particular RNA-RNA interactions. In this updated version, we have added novel tools for RNA secondary structure prediction and have significantly updated the visualisation functionality. The new method CoBold can identify transient RNA structure features and their potential functional effects on a known RNA structure during co-transcriptional structure formation. New tool ShapeSorter can predict evolutionarily conserved RNA secondary structure features while simultaneously taking experimental SHAPE probing evidence into account. The web-server R-Chie which visualises RNA secondary structure information in terms of arc diagrams, can now be used to also visualise and intuitively compare RNA-RNA, RNA-DNA and DNA-DNA interactions alongside multiple sequence alignments and quantitative information. The prediction generated by any method in e-RNA can be readily visualised on the web-server. For completed tasks, users can download their results and readily visualise them later on with R-Chie without having to re-run the predictions. e-RNA can be found at http://www.e-rna.org.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


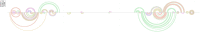
Similar articles
-
e-RNA: a collection of web servers for comparative RNA structure prediction and visualisation.Nucleic Acids Res. 2014 Jul;42(Web Server issue):W373-6. doi: 10.1093/nar/gku292. Epub 2014 May 7. Nucleic Acids Res. 2014. PMID: 24810851 Free PMC article.
-
R-CHIE: a web server and R package for visualizing RNA secondary structures.Nucleic Acids Res. 2012 Jul;40(12):e95. doi: 10.1093/nar/gks241. Epub 2012 Mar 19. Nucleic Acids Res. 2012. PMID: 22434875 Free PMC article.
-
R-chie: a web server and R package for visualizing cis and trans RNA-RNA, RNA-DNA and DNA-DNA interactions.Nucleic Acids Res. 2020 Oct 9;48(18):e105. doi: 10.1093/nar/gkaa708. Nucleic Acids Res. 2020. PMID: 32976561 Free PMC article.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
-
RNA Structure Prediction, Analysis, and Design: An Introduction to Web-Based Tools.Methods Mol Biol. 2022;2518:253-269. doi: 10.1007/978-1-0716-2421-0_15. Methods Mol Biol. 2022. PMID: 35666450 Review.
Cited by
-
RegRNA 3.0: expanding regulatory RNA analysis with new features for motif, interaction, and annotation.Nucleic Acids Res. 2025 Jul 7;53(W1):W485-W495. doi: 10.1093/nar/gkaf405. Nucleic Acids Res. 2025. PMID: 40396374 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

