Viral synergism suppresses R gene-mediated resistance by impairing downstream defense mechanisms in soybean
- PMID: 37099452
- PMCID: PMC10400036
- DOI: 10.1093/plphys/kiad255
Viral synergism suppresses R gene-mediated resistance by impairing downstream defense mechanisms in soybean
Abstract
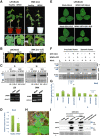
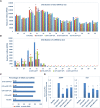
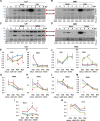
Viral synergism occurs when mixed infection of a susceptible plant by 2 or more viruses leads to increased susceptibility to at least 1 of the viruses. However, the ability of 1 virus to suppress R gene-controlled resistance against another virus has never been reported. In soybean (Glycine max), extreme resistance (ER) against soybean mosaic virus (SMV), governed by the Rsv3 R-protein, manifests a swift asymptomatic resistance against the avirulent strain SMV-G5H. Still, the mechanism by which Rsv3 confers ER is not fully understood. Here, we show that viral synergism broke this resistance by impairing downstream defense mechanisms triggered by Rsv3 activation. We found that activation of the antiviral RNA-silencing pathway and the proimmune mitogen-activated protein kinase 3 (MAPK3), along with the suppression of the proviral MAPK6, are hallmarks of Rsv3-mediated ER against SMV-G5H. Surprisingly, infection with bean pod mottle virus (BPMV) disrupted this ER, allowing SMV-G5H to accumulate in Rsv3-containing plants. BPMV subverted downstream defenses by impairing the RNA-silencing pathway and activating MAPK6. Further, BPMV reduced the accumulation of virus-related siRNAs and increased the virus-activated siRNA that targeted several defense-related nucleotide-binding leucine-rich repeat receptor (NLR) genes through the action of the suppression of RNA-silencing activities encoded in its large and small coat protein subunits. These results illustrate that viral synergism can result from abolishing highly specific R gene resistance by impairing active mechanisms downstream of the R gene.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures




Similar articles
-
Elements Involved in the Rsv3-Mediated Extreme Resistance against an Avirulent Strain of Soybean Mosaic Virus.Viruses. 2018 Oct 24;10(11):581. doi: 10.3390/v10110581. Viruses. 2018. PMID: 30355968 Free PMC article.
-
An Avirulent Strain of Soybean Mosaic Virus Reverses the Defensive Effect of Abscisic Acid in a Susceptible Soybean Cultivar.Viruses. 2019 Sep 19;11(9):879. doi: 10.3390/v11090879. Viruses. 2019. PMID: 31546878 Free PMC article.
-
RNAi-mediated SMV P3 cistron silencing confers significantly enhanced resistance to multiple Potyvirus strains and isolates in transgenic soybean.Plant Cell Rep. 2018 Jan;37(1):103-114. doi: 10.1007/s00299-017-2186-0. Epub 2017 Jul 29. Plant Cell Rep. 2018. PMID: 28756582
-
Isolation and validation of a candidate Rsv3 gene from a soybean genotype that confers strain-specific resistance to soybean mosaic virus.Virology. 2018 Jan 1;513:153-159. doi: 10.1016/j.virol.2017.10.014. Epub 2017 Nov 5. Virology. 2018. PMID: 29080441
-
Transgene Silencing, RNA Interference, and the Antiviral Defense Mechanism Directed by Small Interfering RNAs.Phytopathology. 2023 Apr;113(4):616-625. doi: 10.1094/PHYTO-10-22-0358-IA. Epub 2023 Apr 26. Phytopathology. 2023. PMID: 36441873 Review.
Cited by
-
Exploring sugar allocation and metabolic shifts in cassava plants infected with Cassava common mosaic virus (CsCMV) under long-day photoperiod: diel changes in source and sink leaves.J Plant Res. 2025 Jan;138(1):131-145. doi: 10.1007/s10265-024-01595-4. Epub 2024 Nov 19. J Plant Res. 2025. PMID: 39560817
-
Crop antiviral defense: Past and future perspective.Sci China Life Sci. 2024 Dec;67(12):2617-2634. doi: 10.1007/s11427-024-2680-3. Epub 2024 Aug 15. Sci China Life Sci. 2024. PMID: 39190125 Review.
-
WGCNA Reveals Hub Genes and Key Gene Regulatory Pathways of the Response of Soybean to Infection by Soybean mosaic virus.Genes (Basel). 2024 Apr 27;15(5):566. doi: 10.3390/genes15050566. Genes (Basel). 2024. PMID: 38790195 Free PMC article.
-
A tritrophic plant-insect-pathogen system used to develop a closely linked Rag2 and Rsv1-h recombinant haplotype in double-resistant soybean germplasm.BMC Genomics. 2025 May 28;26(1):539. doi: 10.1186/s12864-025-11686-8. BMC Genomics. 2025. PMID: 40426039 Free PMC article.
-
Deciphering the landscape and evolutionary trajectory of NLR immune receptors in Dioscorea alata.Plant Mol Biol. 2024 Dec 25;115(1):13. doi: 10.1007/s11103-024-01541-x. Plant Mol Biol. 2024. PMID: 39720984
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

