Full-length prion protein incorporated into prion aggregates is a marker for prion strain-specific destabilization of aggregate structure following cellular uptake
- PMID: 37099550
- PMCID: PMC10506170
- DOI: 10.1093/jb/mvad032
Full-length prion protein incorporated into prion aggregates is a marker for prion strain-specific destabilization of aggregate structure following cellular uptake
Abstract
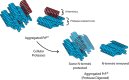
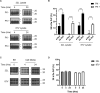
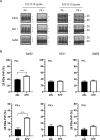
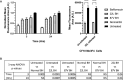
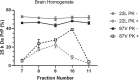
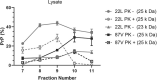
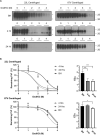
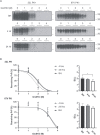
Accumulation of insoluble aggregates of infectious, partially protease-resistant prion protein (PrPD) generated via the misfolding of protease sensitive prion protein (PrPC) into the same infectious conformer, is a hallmark of prion diseases. Aggregated PrPD is taken up and degraded by cells, a process likely involving changes in aggregate structure that can be monitored by accessibility of the N-terminus of full-length PrPD to cellular proteases. We therefore tracked the protease sensitivity of full-length PrPD before and after cellular uptake for two murine prion strains, 22L and 87V. For both strains, PrPD aggregates were less stable following cellular uptake with increased accessibility of the N-terminus to cellular proteases across most aggregate sizes. However, a limited size range of aggregates was able to better protect the N-termini of full-length PrPD, with the N-terminus of 22L-derived PrPD more protected than that of 87V. Interestingly, changes in aggregate structure were associated with minimal changes to the protease-resistant core of PrPD. Our data show that cells destabilize the aggregate quaternary structure protecting PrPD from proteases in a strain-dependent manner, with structural changes exposing protease sensitive PrPD having little effect on the protease-resistant core, and thus conformation, of aggregated PrPD.
Keywords: PrPD; PrPSc; aggregate structure; degradation; prion; protein.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Japanese Biochemical Society. All rights reserved.
Figures

Similar articles
-
The Size and Stability of Infectious Prion Aggregates Fluctuate Dynamically during Cellular Uptake and Disaggregation.Biochemistry. 2021 Feb 9;60(5):398-411. doi: 10.1021/acs.biochem.0c00923. Epub 2021 Jan 26. Biochemistry. 2021. PMID: 33497187
-
A specific population of abnormal prion protein aggregates is preferentially taken up by cells and disaggregated in a strain-dependent manner.J Virol. 2013 Nov;87(21):11552-61. doi: 10.1128/JVI.01484-13. Epub 2013 Aug 21. J Virol. 2013. PMID: 23966386 Free PMC article.
-
Pathogenic mutations within the hydrophobic domain of the prion protein lead to the formation of protease-sensitive prion species with increased lethality.J Virol. 2014 Mar;88(5):2690-703. doi: 10.1128/JVI.02720-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352465 Free PMC article.
-
Properties and pathogenicity of prion-derived peptides.Protein Pept Lett. 2009;16(3):230-8. doi: 10.2174/092986609787601651. Protein Pept Lett. 2009. PMID: 19275735 Review.
-
Prion protein conversion in vitro.J Mol Med (Berl). 2004 Jun;82(6):348-56. doi: 10.1007/s00109-004-0534-3. Epub 2004 Mar 10. J Mol Med (Berl). 2004. PMID: 15014886 Review.
Cited by
-
Chaperone-mediated disaggregation of infectious prions releases particles that seed new prion formation in a strain-specific manner.J Biol Chem. 2025 Jan;301(1):108062. doi: 10.1016/j.jbc.2024.108062. Epub 2024 Dec 9. J Biol Chem. 2025. PMID: 39662829 Free PMC article.
References
-
- Büeler, H., Fischer, M., Lang, Y., Bluethmann, H., Lipp, H.-P., DeArmond, S.J., Prusiner, S.B., Aguet, M., and Weissmann, C. (1992) Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 356, 577–582 - PubMed
-
- Gabus, C., Derrington, E., Leblanc, P., Chnaiderman, J., Dormont, D., Swietnicki, W., Morillas, M., Surewicz, W.K., Marc, D., Nandi, P., and Darlix, J.L. (2001) The prion protein has RNA binding and chaperoning properties characteristic of nucleocapsid protein NCP7 of HIV-1. J. Biol. Chem. 276, 19301–19309 - PubMed
-
- Bremer, J., Baumann, F., Tiberi, C., Wessig, C., Fischer, H., Schwarz, P., Steele, A.D., Toyka, K.V., Nave, K.-A., Weis, J., and Aguzzi, A. (2010) Axonal prion protein is required for peripheral myelin maintenance. Nat. Neurosci. 13, 310–318 - PubMed

