Sleep deprivation exacerbates microglial reactivity and Aβ deposition in a TREM2-dependent manner in mice
- PMID: 37099634
- PMCID: PMC10449561
- DOI: 10.1126/scitranslmed.ade6285
Sleep deprivation exacerbates microglial reactivity and Aβ deposition in a TREM2-dependent manner in mice
Abstract
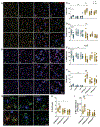
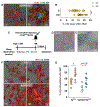
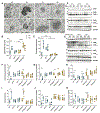
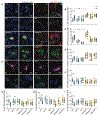
Sleep loss is associated with cognitive decline in the aging population and is a risk factor for Alzheimer's disease (AD). Considering the crucial role of immunomodulating genes such as that encoding the triggering receptor expressed on myeloid cells type 2 (TREM2) in removing pathogenic amyloid-β (Aβ) plaques and regulating neurodegeneration in the brain, our aim was to investigate whether and how sleep loss influences microglial function in mice. We chronically sleep-deprived wild-type mice and the 5xFAD mouse model of cerebral amyloidosis, expressing either the humanized TREM2 common variant, the loss-of-function R47H AD-associated risk variant, or without TREM2 expression. Sleep deprivation not only enhanced TREM2-dependent Aβ plaque deposition compared with 5xFAD mice with normal sleeping patterns but also induced microglial reactivity that was independent of the presence of parenchymal Aβ plaques. We investigated lysosomal morphology using transmission electron microscopy and found abnormalities particularly in mice without Aβ plaques and also observed lysosomal maturation impairments in a TREM2-dependent manner in both microglia and neurons, suggesting that changes in sleep modified neuro-immune cross-talk. Unbiased transcriptome and proteome profiling provided mechanistic insights into functional pathways triggered by sleep deprivation that were unique to TREM2 and Aβ pathology and that converged on metabolic dyshomeostasis. Our findings highlight that sleep deprivation directly affects microglial reactivity, for which TREM2 is required, by altering the metabolic ability to cope with the energy demands of prolonged wakefulness, leading to further Aβ deposition, and underlines the importance of sleep modulation as a promising future therapeutic approach.
Conflict of interest statement
Competing interests
DMH is as an inventor on a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies, US patent 9,834,596. DMH co-founded and is on the scientific advisory board of C2N Diagnostics. DMH is on the scientific advisory board of Denali, Genentech, and Cajal Neuroscience and consults for Alector and Asteroid
Figures


References
-
- Achariyar TM, Li B, Peng W, Verghese PB, Shi Y, McConnell E, Benraiss A, Kasper T, Song W, Takano T, Holtzman DM, Nedergaard M, Deane R, Glymphatic distribution of CSF-derived apoE into brain is isoform specific and suppressed during sleep deprivation. Mol. Neurodegener 11, 74 (2016). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

