Integrative genetic analysis identifies FLVCR1 as a plasma-membrane choline transporter in mammals
- PMID: 37100056
- PMCID: PMC10367582
- DOI: 10.1016/j.cmet.2023.04.003
Integrative genetic analysis identifies FLVCR1 as a plasma-membrane choline transporter in mammals
Abstract
Genome-wide association studies (GWASs) of serum metabolites have the potential to uncover genes that influence human metabolism. Here, we combined an integrative genetic analysis that associates serum metabolites to membrane transporters with a coessentiality map of metabolic genes. This analysis revealed a connection between feline leukemia virus subgroup C cellular receptor 1 (FLVCR1) and phosphocholine, a downstream metabolite of choline metabolism. Loss of FLVCR1 in human cells strongly impairs choline metabolism due to the inhibition of choline import. Consistently, CRISPR-based genetic screens identified phospholipid synthesis and salvage machinery as synthetic lethal with FLVCR1 loss. Cells and mice lacking FLVCR1 exhibit structural defects in mitochondria and upregulate integrated stress response (ISR) through heme-regulated inhibitor (HRI) kinase. Finally, Flvcr1 knockout mice are embryonic lethal, which is partially rescued by choline supplementation. Altogether, our findings propose FLVCR1 as a major choline transporter in mammals and provide a platform to discover substrates for unknown metabolite transporters.
Keywords: FLVCR1; PCARP; choline; metabolism; mitochondria; phosphatidylcholine.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.B. is scientific advisor to Nanocare Pharmaceuticals and Atavistik Bio.
Figures


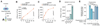


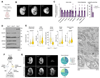
Comment in
-
The cellular supply-side economics for phospholipids.Cell Metab. 2023 Jun 6;35(6):909-911. doi: 10.1016/j.cmet.2023.05.004. Cell Metab. 2023. PMID: 37285806
References
-
- Neal B., Perkovic V., Mahaffey KW., de Zeeuw D., Fulcher G., Erondu N., Shaw W., Law G., Desai M., and Matthews DR. (2017). Canagliflozin and Cardiovascular and Renal Events in Type 2 Diabetes. New England Journal of Medicine 377, 644–657. 10.1056/NEJMOA1611925/SUPPL_FILE/NEJMOA1611925_DISCLOSURES.PDF. - DOI - PubMed
-
- Mann JJ (2005). The Medical Management of Depression. - PubMed
-
- Yin X, Chan LS, Bose D, Jackson AU, VandeHaar P, Locke AE, Fuchsberger C, Stringham HM, Welch R, Yu K, et al. (2022). Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nature Communications 2022 13:1 13, 1–14. 10.1038/s41467-022-29143-5. - DOI - PMC - PubMed
-
- Goldstein JL, and Brown MS (1974). Binding and Degradation of Low Density Lipoproteins by Cultured Human Fibroblasts: COMPARISON OF CELLS FROM A NORMAL SUBJECT AND FROM A PATIENT WITH HOMOZYGOUS FAMILIAL HYPERCHOLESTEROLEMIA. Journal of Biological Chemistry 249, 5153–5162. 10.1016/S0021-9258(19)42341-7. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

