Structural insights into ubiquitin chain cleavage by Legionella ovarian tumor deubiquitinases
- PMID: 37100438
- PMCID: PMC10133868
- DOI: 10.26508/lsa.202201876
Structural insights into ubiquitin chain cleavage by Legionella ovarian tumor deubiquitinases
Abstract
Although ubiquitin is found only in eukaryotes, several pathogenic bacteria and viruses possess proteins that hinder the host ubiquitin system. Legionella, a gram-negative intracellular bacterium, possesses an ovarian tumor (OTU) family of deubiquitinases (Lot DUBs). Herein, we describe the molecular characteristics of Lot DUBs. We elucidated the structure of the LotA OTU1 domain and revealed that entire Lot DUBs possess a characteristic extended helical lobe that is not found in other OTU-DUBs. The structural topology of an extended helical lobe is the same throughout the Lot family, and it provides an S1' ubiquitin-binding site. Moreover, the catalytic triads of Lot DUBs resemble those of the A20-type OTU-DUBs. Furthermore, we revealed a unique mechanism by which LotA OTU domains cooperate together to distinguish the length of the chain and preferentially cleave longer K48-linked polyubiquitin chains. The LotA OTU1 domain itself cleaves K6-linked ubiquitin chains, whereas it is also essential for assisting the cleavage of longer K48-linked polyubiquitin chains by the OTU2 domain. Thus, this study provides novel insights into the structure and mechanism of action of Lot DUBs.
© 2023 Kang et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
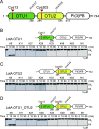









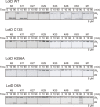


References
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68: 352–367. 10.1107/S0907444912001308 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
