MYT1L is required for suppressing earlier neuronal development programs in the adult mouse brain
- PMID: 37100461
- PMCID: PMC10234307
- DOI: 10.1101/gr.277413.122
MYT1L is required for suppressing earlier neuronal development programs in the adult mouse brain
Abstract
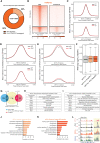
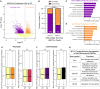
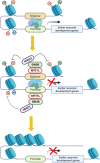
In vitro studies indicate the neurodevelopmental disorder gene myelin transcription factor 1-like (MYT1L) suppresses non-neuronal lineage genes during fibroblast-to-neuron direct differentiation. However, MYT1L's molecular and cellular functions in the adult mammalian brain have not been fully characterized. Here, we found that MYT1L loss leads to up-regulated deep layer (DL) gene expression, corresponding to an increased ratio of DL/UL neurons in the adult mouse cortex. To define potential mechanisms, we conducted Cleavage Under Targets & Release Using Nuclease (CUT&RUN) to map MYT1L binding targets and epigenetic changes following MYT1L loss in mouse developing cortex and adult prefrontal cortex (PFC). We found MYT1L mainly binds to open chromatin, but with different transcription factor co-occupancies between promoters and enhancers. Likewise, multiomic data set integration revealed that, at promoters, MYT1L loss does not change chromatin accessibility but increases H3K4me3 and H3K27ac, activating both a subset of earlier neuronal development genes as well as Bcl11b, a key regulator for DL neuron development. Meanwhile, we discovered that MYT1L normally represses the activity of neurogenic enhancers associated with neuronal migration and neuronal projection development by closing chromatin structures and promoting removal of active histone marks. Further, we showed that MYT1L interacts with HDAC2 and transcriptional repressor SIN3B in vivo, providing potential mechanisms underlying repressive effects on histone acetylation and gene expression. Overall, our findings provide a comprehensive map of MYT1L binding in vivo and mechanistic insights into how MYT1L loss leads to aberrant activation of earlier neuronal development programs in the adult mouse brain.
© 2023 Chen et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Bainor AJ, Saini S, Calderon A, Casado-Polanco R, Giner-Ramirez B, Moncada C, Cantor DJ, Ernlund A, Litovchick L, David G. 2018. The HDAC-associated Sin3B protein represses DREAM complex targets and cooperates with APC/C to promote quiescence. Cell Rep 25: 2797–2807.e8. 10.1016/j.celrep.2018.11.024 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
