Molecular mimicry and cancer vaccine development
- PMID: 37101139
- PMCID: PMC10131527
- DOI: 10.1186/s12943-023-01776-0
Molecular mimicry and cancer vaccine development
Abstract
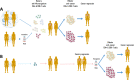
Background: The development of cancer immunotherapeutic strategies relies on the identification and validation of optimal target tumor antigens, which should be tumor-specific as well as able to elicit a swift and potent anti-tumor immune response. The vast majority of such strategies are based on tumor associated antigens (TAAs) which are shared wild type cellular self-epitopes highly expressed on tumor cells. Indeed, TAAs can be used to develop off-the-shelf cancer vaccines appropriate to all patients affected by the same malignancy. However, given that they may be also presented by HLAs on the surface of non-malignant cells, they may be possibly affected by immunological tolerance or elicit autoimmune responses.
Main body: In order to overcome such limitations, analogue peptides with improved antigenicity and immunogenicity able to elicit a cross-reactive T cell response are needed. To this aim, non-self-antigens derived from microorganisms (MoAs) may be of great benefit.
Keywords: Cancer Vaccines; Microbiota; Molecular Mimicry; T cell cross-reactivity; Tumor-Associated Antigens.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures
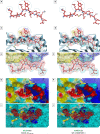
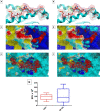
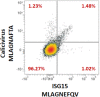
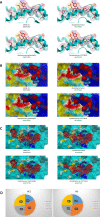
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

