Inulin diet uncovers complex diet-microbiota-immune cell interactions remodeling the gut epithelium
- PMID: 37101209
- PMCID: PMC10131329
- DOI: 10.1186/s40168-023-01520-2
Inulin diet uncovers complex diet-microbiota-immune cell interactions remodeling the gut epithelium
Erratum in
-
Correction: Inulin diet uncovers complex diet-microbiota-immune cell interactions remodeling the gut epithelium.Microbiome. 2023 May 31;11(1):122. doi: 10.1186/s40168-023-01579-x. Microbiome. 2023. PMID: 37259114 Free PMC article. No abstract available.
Abstract
Background: The continuous proliferation of intestinal stem cells followed by their tightly regulated differentiation to epithelial cells is essential for the maintenance of the gut epithelial barrier and its functions. How these processes are tuned by diet and gut microbiome is an important, but poorly understood question. Dietary soluble fibers, such as inulin, are known for their ability to impact the gut bacterial community and gut epithelium, and their consumption has been usually associated with health improvement in mice and humans. In this study, we tested the hypothesis that inulin consumption modifies the composition of colonic bacteria and this impacts intestinal stem cells functions, thus affecting the epithelial structure.
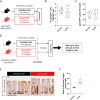
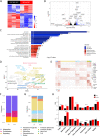
Methods: Mice were fed with a diet containing 5% of the insoluble fiber cellulose or the same diet enriched with an additional 10% of inulin. Using a combination of histochemistry, host cell transcriptomics, 16S microbiome analysis, germ-free, gnotobiotic, and genetically modified mouse models, we analyzed the impact of inulin intake on the colonic epithelium, intestinal bacteria, and the local immune compartment.
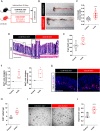
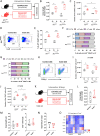
Results: We show that the consumption of inulin diet alters the colon epithelium by increasing the proliferation of intestinal stem cells, leading to deeper crypts and longer colons. This effect was dependent on the inulin-altered gut microbiota, as no modulations were observed in animals deprived of microbiota, nor in mice fed cellulose-enriched diets. We also describe the pivotal role of γδ T lymphocytes and IL-22 in this microenvironment, as the inulin diet failed to induce epithelium remodeling in mice lacking this T cell population or cytokine, highlighting their importance in the diet-microbiota-epithelium-immune system crosstalk.
Conclusion: This study indicates that the intake of inulin affects the activity of intestinal stem cells and drives a homeostatic remodeling of the colon epithelium, an effect that requires the gut microbiota, γδ T cells, and the presence of IL-22. Our study indicates complex cross kingdom and cross cell type interactions involved in the adaptation of the colon epithelium to the luminal environment in steady state. Video Abstract.
Keywords: Bacteroidales; Epithelial remodeling; Gut homeostasis; High-fiber diet; IL-22; Intestinal stem cells; γδ T cells.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Gehart H, Clevers H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol. 2019;16:19–34. - PubMed
-
- Allaire JM, Crowley SM, Law HT, Chang S-Y, Ko H-J, Vallance BA. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018;39:677–696. - PubMed
-
- Xing PY, Pettersson S, Kundu P. Microbial Metabolites and Intestinal Stem Cells Tune Intestinal Homeostasis. Proteomics. 2020;20:1800419. - PubMed
-
- Calibasi-Kocal G, Mashinchian O, Basbinar Y, Ellidokuz E, Cheng C-W, Yilmaz ÖH. Nutritional Control of Intestinal Stem Cells in Homeostasis and Tumorigenesis. Trends Endocrinol Metab. 2021;32:20–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

