Multi-omics reveals Dengzhan Shengmai formulation ameliorates cognitive impairments in D-galactose-induced aging mouse model by regulating CXCL12/CXCR4 and gut microbiota
- PMID: 37101548
- PMCID: PMC10123283
- DOI: 10.3389/fphar.2023.1175970
Multi-omics reveals Dengzhan Shengmai formulation ameliorates cognitive impairments in D-galactose-induced aging mouse model by regulating CXCL12/CXCR4 and gut microbiota
Abstract
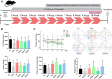
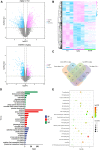
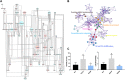
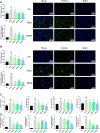
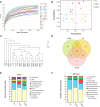
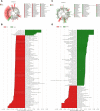
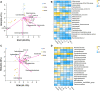
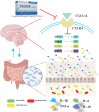
Dengzhan Shengmai (DZSM), a traditional Chinese medicine formulation, has been administered extensively to elderly individuals with cognitive impairment (CI). However, the underlying mechanisms by which Dengzhan Shengmai improves cognitive impairment remains unknown. This study aimed to elucidate the underlying mechanism of the effect of Dengzhan Shengmai on aging-associated cognitive impairment via a comprehensive combination of transcriptomics and microbiota assessment. Dengzhan Shengmai was orally administered to a D-galactose-induced aging mouse model, and evaluation with an open field task (OFT), Morris water maze (MWM), and histopathological staining was performed. Transcriptomics and 16S rDNA sequencing were applied to elucidate the mechanism of Dengzhan Shengmai in alleviating cognitive deficits, and enzyme-linked immunosorbent assay (ELISA), quantitative real-time polymerase chain reaction (PCR), and immunofluorescence were employed to verify the results. The results first confirmed the therapeutic effects of Dengzhan Shengmai against cognitive defects; specifically, Dengzhan Shengmai improved learning and impairment, suppressed neuro loss, and increased Nissl body morphology repair. Comprehensive integrated transcriptomics and microbiota analysis indicated that chemokine CXC motif receptor 4 (CXCR4) and its ligand CXC chemokine ligand 12 (CXCL12) were targets for improving cognitive impairments with Dengzhan Shengmai and also indirectly suppressed the intestinal flora composition. Furthermore, in vivo results confirmed that Dengzhan Shengmai suppressed the expression of CXC motif receptor 4, CXC chemokine ligand 12, and inflammatory cytokines. This suggested that Dengzhan Shengmai inhibited CXC chemokine ligand 12/CXC motif receptor 4 expression and modulated intestinal microbiome composition by influencing inflammatory factors. Thus, Dengzhan Shengmai improves aging-related cognitive impairment effects via decreased CXC chemokine ligand 12/CXC motif receptor 4 and inflammatory factor modulation to improve gut microbiota composition.
Keywords: CXCL12; CXCR4; Dengzhan Shengmai (DZSM); aging; cognitive impairment (CI).
Copyright © 2023 Hou, Xu, Cao, Tian, Wang, Zhu, Zhang and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Cattaneo A., Cattane N., Galluzzi S., Provasi S., Lopizzo N., Festari C., et al. (2017). Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 49, 60–68. 10.1016/j.neurobiolaging.2016.08.019 - DOI - PubMed
LinkOut - more resources
Full Text Sources

