Targeting Oncogene Promoters and Ribosomal RNA Biogenesis by G-Quadruplex Binding Ligands Translate to Anticancer Activity
- PMID: 37101746
- PMCID: PMC10114666
- DOI: 10.1021/acsbiomedchemau.1c00039
Targeting Oncogene Promoters and Ribosomal RNA Biogenesis by G-Quadruplex Binding Ligands Translate to Anticancer Activity
Abstract
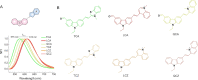
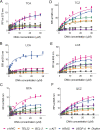
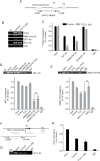
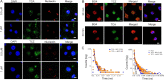
G-Quadruplex (GQ) nucleic acids are promising therapeutic targets in anticancer research due to their structural robustness, polymorphism, and gene-regulatory functions. Here, we presented the structure-activity relationship of carbazole-based monocyanine ligands using region-specific functionalization with benzothiazole (TCA and TCZ), lepidine (LCA and LCZ), and quinaldine (QCA and QCZ) acceptor moieties and evaluated their binding profiles with different oncogenic GQs. Their differential turn-on fluorescence emission upon GQ binding confirmed the GQ-to-duplex selectivity of all carbazole ligands, while the isothermal titration calorimetry results showed selective interactions of TCZ and TCA to c-MYC and BCL-2 GQs, respectively. The aldehyde group in TCA favors stacking interactions with the tetrad of BCL-2 GQ, whereas TCZ provides selective groove interactions with c-MYC GQ. Dual-luciferase assay and chromatin immunoprecipitation (ChIP) showed that these molecules interfere with the recruitment of specific transcription factors at c-MYC and BCL-2 promoters and stabilize the promoter GQ structures to inhibit their constitutive transcription in cancer cells. Their intrinsic turn-on fluorescence response with longer lifetimes upon GQ binding allowed real-time visualization of GQ structures at subcellular compartments. Confocal microscopy revealed the uptake of these ligands in the nucleoli, resulting in nucleolar stress. ChIP studies further confirmed the inhibition of Nucleolin occupancy at multiple GQ-enriched regions of ribosomal DNA (rDNA) promoters, which arrested rRNA biogenesis. Therefore, carbazole ligands act as the "double-edged swords" to arrest c-MYC and BCL-2 overexpression as well as rRNA biogenesis, triggering synergistic inhibition of multiple oncogenic pathways and apoptosis in cancer cells.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Neidle S.; Balasubramanian S., Quadruplex nucleic acids; Royal Society of Chemistry: 2007; Vol. 7.
-
- Drygin D.; Siddiqui-Jain A.; O’Brien S.; Schwaebe M.; Lin A.; Bliesath J.; Ho C. B.; Proffitt C.; Trent K.; Whitten J. P.; Lim J. K. C.; von Hoff D.; Anderes K.; Rice W. G. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. 10.1158/0008-5472.CAN-09-1304. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
