Six-Transmembrane Epithelial Antigen of Prostate 4: An Indicator of Prognosis and Tumor Immunity in Hepatocellular Carcinoma
- PMID: 37101765
- PMCID: PMC10124562
- DOI: 10.2147/JHC.S394973
Six-Transmembrane Epithelial Antigen of Prostate 4: An Indicator of Prognosis and Tumor Immunity in Hepatocellular Carcinoma
Abstract
Purpose: The six-transmembrane epithelial antigen of prostate 4 (STEAP4) has been linked to tumor progression via its involvement in inflammatory responses, oxidative stress, and metabolism. However, STEAP4 has rarely been studied in hepatocellular carcinoma (HCC). We explored STEAP4 expression associated with tumor prognosis to understand its role in tumor biology in HCC.
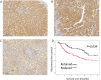
Patients and methods: STEAP4 mRNA and protein expressions were primarily analyzed using bioinformatics tools based on The Cancer Genome Atlas database to understand the expression pattern, molecular mechanism, prognostic impact, and association with immune cell infiltration. We further investigated the association between STEAP4 protein expression and clinicopathological parameters and their predictive value in HCC patients using immunohistochemical staining of tissue microarrays.
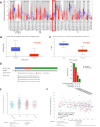
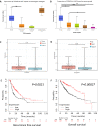
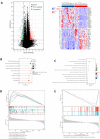
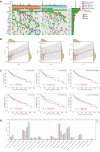
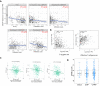
Results: The expression of STEAP4 mRNA and protein in HCC tissues was significantly lower than in normal liver tissues. Reduced expression of STEAP4 was linked to advanced HCC stages, poor recurrence-free survival (RFS), and overall survival. Furthermore, reduced STEAP4 expression was a significant predictor of worse RFS in univariate and multivariate analyses in the immunohistochemical cohort. GO, KEGG, and GSEA analyses revealed that STEAP4 is related to numerous biological processes and pathways, including drug metabolism, DNA replication, RNA metabolism, and immune response. In terms of the immune system, the decreased level of STEAP4 was correlated with the immunosuppressive microenvironment.
Conclusion: Our data indicated that reduced STEAP4 expression was significantly associated with tumor aggressiveness and poor prognosis, possibly because of its link to various biological processes and induction of HCC immune evasion. Therefore, STEAP4 expression may serve as a potential prognostic biomarker for cancer progression and immunity, as well as a therapeutic target in HCC.
Keywords: Hepatocellular carcinoma; bioinformatics; immunohistochemistry; prognostic factor.
© 2023 Ju et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures
References
-
- Müller M, Bird TG, Nault JC. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol. 2020;72:990–1002. - PubMed
LinkOut - more resources
Full Text Sources

