Artificial intelligence-enabled classification of hypertrophic heart diseases using electrocardiograms
- PMID: 37101945
- PMCID: PMC10123506
- DOI: 10.1016/j.cvdhj.2023.03.001
Artificial intelligence-enabled classification of hypertrophic heart diseases using electrocardiograms
Abstract
Background: Differentiating among cardiac diseases associated with left ventricular hypertrophy (LVH) informs diagnosis and clinical care.
Objective: To evaluate if artificial intelligence-enabled analysis of the 12-lead electrocardiogram (ECG) facilitates automated detection and classification of LVH.
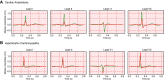
Methods: We used a pretrained convolutional neural network to derive numerical representations of 12-lead ECG waveforms from patients in a multi-institutional healthcare system who had cardiac diseases associated with LVH (n = 50,709), including cardiac amyloidosis (n = 304), hypertrophic cardiomyopathy (n = 1056), hypertension (n = 20,802), aortic stenosis (n = 446), and other causes (n = 4766). We then regressed LVH etiologies relative to no LVH on age, sex, and the numerical 12-lead representations using logistic regression ("LVH-Net"). To assess deep learning model performance on single-lead data analogous to mobile ECGs, we also developed 2 single-lead deep learning models by training models on lead I ("LVH-Net Lead I") or lead II ("LVH-Net Lead II") from the 12-lead ECG. We compared the performance of the LVH-Net models to alternative models fit on (1) age, sex, and standard ECG measures, and (2) clinical ECG-based rules for diagnosing LVH.
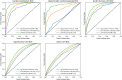
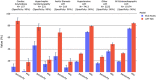
Results: The areas under the receiver operator characteristic curve of LVH-Net by specific LVH etiology were cardiac amyloidosis 0.95 [95% CI, 0.93-0.97], hypertrophic cardiomyopathy 0.92 [95% CI, 0.90-0.94], aortic stenosis LVH 0.90 [95% CI, 0.88-0.92], hypertensive LVH 0.76 [95% CI, 0.76-0.77], and other LVH 0.69 [95% CI 0.68-0.71]. The single-lead models also discriminated LVH etiologies well.
Conclusion: An artificial intelligence-enabled ECG model is favorable for detection and classification of LVH and outperforms clinical ECG-based rules.
Keywords: Artificial intelligence; Cardiac amyloidosis; Electrocardiography; Hypertrophic cardiomyopathy; Hypertrophic heart disease.
© 2023 Pub-.
Figures



References
-
- Vakili B.A., Okin P.M., Devereux R.B. Prognostic implications of left ventricular hypertrophy. Am Heart J. 2001;141:334–341. - PubMed
-
- Savage D.D., Garrison R.J., Kannel W.B., et al. The spectrum of left ventricular hypertrophy in a general population sample: the Framingham Study. Circulation. 1987;75:I26–I33. - PubMed
-
- Gertz M.A., Dispenzieri A. Systemic amyloidosis recognition, prognosis, and therapy: a systematic review. JAMA. 2020;324:79–89. - PubMed
-
- Gradman A.H., Alfayoumi F. From left ventricular hypertrophy to congestive heart failure: management of hypertensive heart disease. Prog Cardiovasc Dis. 2006;48:326–341. - PubMed
-
- Makavos G., Kappaairis C., Tselegkidi M.E., et al. Hypertrophic cardiomyopathy: an updated review on diagnosis, prognosis, and treatment. Heart Fail Rev. 2019;24:439–459. - PubMed
LinkOut - more resources
Full Text Sources

