Data-Driven Elucidation of Flavor Chemistry
- PMID: 37102791
- PMCID: PMC10176570
- DOI: 10.1021/acs.jafc.3c00909
Data-Driven Elucidation of Flavor Chemistry
Abstract
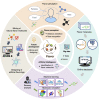
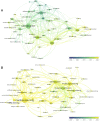
Flavor molecules are commonly used in the food industry to enhance product quality and consumer experiences but are associated with potential human health risks, highlighting the need for safer alternatives. To address these health-associated challenges and promote reasonable application, several databases for flavor molecules have been constructed. However, no existing studies have comprehensively summarized these data resources according to quality, focused fields, and potential gaps. Here, we systematically summarized 25 flavor molecule databases published within the last 20 years and revealed that data inaccessibility, untimely updates, and nonstandard flavor descriptions are the main limitations of current studies. We examined the development of computational approaches (e.g., machine learning and molecular simulation) for the identification of novel flavor molecules and discussed their major challenges regarding throughput, model interpretability, and the lack of gold-standard data sets for equitable model evaluation. Additionally, we discussed future strategies for the mining and designing of novel flavor molecules based on multi-omics and artificial intelligence to provide a new foundation for flavor science research.
Keywords: active ingredients; bioinformatics; cheminformatics; database; machine learning.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Artificial intelligence and food flavor: How AI models are shaping the future and revolutionary technologies for flavor food development.Compr Rev Food Sci Food Saf. 2025 Jan;24(1):e70068. doi: 10.1111/1541-4337.70068. Compr Rev Food Sci Food Saf. 2025. PMID: 39783879 Review.
-
FlavorDB: a database of flavor molecules.Nucleic Acids Res. 2018 Jan 4;46(D1):D1210-D1216. doi: 10.1093/nar/gkx957. Nucleic Acids Res. 2018. PMID: 29059383 Free PMC article.
-
Rethinking Drug Repositioning and Development with Artificial Intelligence, Machine Learning, and Omics.OMICS. 2019 Nov;23(11):539-548. doi: 10.1089/omi.2019.0151. Epub 2019 Oct 25. OMICS. 2019. PMID: 31651216
-
A review on utilizing machine learning technology in the fields of electronic emergency triage and patient priority systems in telemedicine: Coherent taxonomy, motivations, open research challenges and recommendations for intelligent future work.Comput Methods Programs Biomed. 2021 Sep;209:106357. doi: 10.1016/j.cmpb.2021.106357. Epub 2021 Aug 16. Comput Methods Programs Biomed. 2021. PMID: 34438223 Review.
-
Demystifying food flavor: Flavor data interpretation through machine learning.Food Chem. 2025 Aug 15;483:144000. doi: 10.1016/j.foodchem.2025.144000. Epub 2025 Mar 26. Food Chem. 2025. PMID: 40233512
Cited by
-
Relevance of Phytochemical Taste for Anti-Cancer Activity: A Statistical Inquiry.Int J Mol Sci. 2023 Nov 12;24(22):16227. doi: 10.3390/ijms242216227. Int J Mol Sci. 2023. PMID: 38003415 Free PMC article. Review.
-
Discovery of potential anti- Staphylococcus aureus natural products and their mechanistic studies using machine learning and molecular dynamic simulations.Heliyon. 2024 Apr 26;10(9):e30389. doi: 10.1016/j.heliyon.2024.e30389. eCollection 2024 May 15. Heliyon. 2024. PMID: 38737232 Free PMC article.
-
Development of a Mixed Solution Certified Reference Material of Vanillin Spices.ACS Omega. 2025 Jun 1;10(22):23601-23608. doi: 10.1021/acsomega.5c02463. eCollection 2025 Jun 10. ACS Omega. 2025. PMID: 40521479 Free PMC article.
-
Research on Bitter Peptides in the Field of Bioinformatics: A Comprehensive Review.Int J Mol Sci. 2024 Sep 12;25(18):9844. doi: 10.3390/ijms25189844. Int J Mol Sci. 2024. PMID: 39337334 Free PMC article. Review.
-
Mlp4green: A Binary Classification Approach Specifically for Green Odor.Int J Mol Sci. 2024 Mar 20;25(6):3515. doi: 10.3390/ijms25063515. Int J Mol Sci. 2024. PMID: 38542486 Free PMC article.
References
-
- Eelager M. P.; Masti S. P.; Chougale R. B.; Hiremani V. D.; Narasgoudar S. S.; Dalbanjan N. P.; S.K. P. K. Evaluation of mechanical, antimicrobial, and antioxidant properties of vanillic acid induced chitosan/poly (vinyl alcohol) active films to prolong the shelf life of green chilli. Int. J. Biol. Macromol. 2023, 232, 123499.10.1016/j.ijbiomac.2023.123499. - DOI - PubMed
-
- Zhang D.; Ouyang S.; Cai M.; Zhang H.; Ding S.; Liu D.; Cai P.; Le Y.; Hu Q. N. FADB-China: A molecular-level food adulteration database in China based on molecular fingerprints and similarity algorithms prediction expansion. Food Chem. 2020, 327, 127010.10.1016/j.foodchem.2020.127010. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

