Role of the NaHCO3 Transporter MpsABC in the NaHCO3-β-Lactam-Responsive Phenotype in Methicillin-Resistant Staphylococcus aureus
- PMID: 37102972
- PMCID: PMC10269494
- DOI: 10.1128/spectrum.00141-23
Role of the NaHCO3 Transporter MpsABC in the NaHCO3-β-Lactam-Responsive Phenotype in Methicillin-Resistant Staphylococcus aureus
Abstract
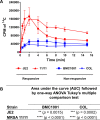
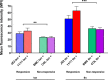
Methicillin-resistant Staphylococcus aureus (MRSA) infections are an increasing concern due to their intrinsic resistance to most standard-of-care β-lactam antibiotics. Recent studies of clinical isolates have documented a novel phenotype, termed NaHCO3 responsiveness, in which a substantial proportion of MRSA strains exhibit enhanced susceptibility to β-lactams such as cefazolin and oxacillin in the presence of NaHCO3. A bicarbonate transporter, MpsAB (
Keywords: CO2; MRSA; MpsABC; bicarbonate transporter; membrane potential-generating system ABC; methicillin-resistant Staphylococcus aureus; sodium bicarbonate; β-lactams.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Bicarbonate Within: A Hidden Modulator of Antibiotic Susceptibility.Antibiotics (Basel). 2025 Jan 16;14(1):96. doi: 10.3390/antibiotics14010096. Antibiotics (Basel). 2025. PMID: 39858381 Free PMC article. Review.
-
Influence of Sodium Bicarbonate on Wall Teichoic Acid Synthesis and β-Lactam Sensitization in NaHCO3-Responsive and Nonresponsive Methicillin-Resistant Staphylococcus aureus.Microbiol Spectr. 2022 Dec 21;10(6):e0342222. doi: 10.1128/spectrum.03422-22. Epub 2022 Nov 15. Microbiol Spectr. 2022. PMID: 36377886 Free PMC article.
-
Genomic investigation and clinical correlates of the in vitro β-lactam: NaHCO3 responsiveness phenotype among methicillin-resistant Staphylococcus aureus isolates from a randomized clinical trial.Antimicrob Agents Chemother. 2024 Jul 9;68(7):e0021824. doi: 10.1128/aac.00218-24. Epub 2024 Jun 5. Antimicrob Agents Chemother. 2024. PMID: 38837393 Free PMC article. Clinical Trial.
-
Bicarbonate Resensitization of Methicillin-Resistant Staphylococcus aureus to β-Lactam Antibiotics.Antimicrob Agents Chemother. 2019 Jun 24;63(7):e00496-19. doi: 10.1128/AAC.00496-19. Print 2019 Jul. Antimicrob Agents Chemother. 2019. PMID: 31010857 Free PMC article.
-
Measuring beta-lactam minimum inhibitory concentrations in Staphylococcus aureus in the clinical microbiology laboratory: pinning the tail on the donkey.J Clin Microbiol. 2024 Jan 17;62(1):e0036623. doi: 10.1128/jcm.00366-23. Epub 2023 Nov 15. J Clin Microbiol. 2024. PMID: 37966224 Free PMC article. Review.
Cited by
-
Staphylococcus aureus Stress Response to Bicarbonate Depletion.Int J Mol Sci. 2024 Aug 26;25(17):9251. doi: 10.3390/ijms25179251. Int J Mol Sci. 2024. PMID: 39273203 Free PMC article.
-
Phenotypic and genotypic correlates of the sodium bicarbonate-responsive phenotype among methicillin-resistant Staphylococcus aureus isolates from skin and soft-tissue infections.Clin Microbiol Infect. 2025 Apr;31(4):588-593. doi: 10.1016/j.cmi.2024.11.034. Epub 2024 Dec 1. Clin Microbiol Infect. 2025. PMID: 39626860
-
Bicarbonate Within: A Hidden Modulator of Antibiotic Susceptibility.Antibiotics (Basel). 2025 Jan 16;14(1):96. doi: 10.3390/antibiotics14010096. Antibiotics (Basel). 2025. PMID: 39858381 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

