Genetic basis for probiotic yeast phenotypes revealed by nanopore sequencing
- PMID: 37103477
- PMCID: PMC10411601
- DOI: 10.1093/g3journal/jkad093
Genetic basis for probiotic yeast phenotypes revealed by nanopore sequencing
Abstract
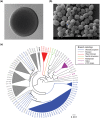
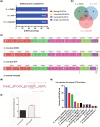
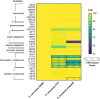
Probiotic yeasts are emerging as preventative and therapeutic solutions for disease. Often ingested via cultured foods and beverages, they can survive the harsh conditions of the gastrointestinal tract and adhere to it, where they provide nutrients and inhibit pathogens like Candida albicans. Yet, little is known of the genomic determinants of these beneficial traits. To this end, we have sequenced 2 food-derived probiotic yeast isolates that mitigate fungal infections. We find that the first strain, KTP, is a strain of Saccharomyces cerevisiae within a small clade that lacks any apparent ancestry from common European/wine S. cerevisiae strains. Significantly, we show that S. cerevisiae KTP genes involved in general stress, pH tolerance, and adherence are markedly different from S. cerevisiae S288C but are similar to the commercial probiotic yeast species S. boulardii. This suggests that even though S. cerevisiae KTP and S. boulardii are from different clades, they may achieve probiotic effect through similar genetic mechanisms. We find that the second strain, ApC, is a strain of Issatchenkia occidentalis, one of the few of this family of yeasts to be sequenced. Because of the dissimilarity of its genome structure and gene organization, we infer that I. occidentalis ApC likely achieves a probiotic effect through a different mechanism than the Saccharomyces strains. Therefore, this work establishes a strong genetic link among probiotic Saccharomycetes, advances the genomics of Issatchenkia yeasts, and indicates that probiotic activity is not monophyletic and complimentary mixtures of probiotics could enhance health benefits beyond a single species.
Keywords: FLO genes; Issatchenkia occidentalis strain ApC; Saccharomyces cerevisiae strain KTP; probiotic traits associated genes.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflict of interest.
Figures

References
-
- Ansari JM, Colasacco C, Emmanouil E, Kohlhepp S, Harriott O. Strain-level diversity of commercial probiotic isolates of Bacillus, Lactobacillus, and Saccharomyces species illustrated by molecular identification and phenotypic profiling. PLoS One. 2019;14(3):e0213841. doi: 10.1371/journal.pone.0213841. - DOI - PMC - PubMed
-
- Auesukaree C, Koedrith P, Saenpayavai P, Asvarak T, Benjaphokee S, Sugiyama M, Kaneko Y, Harashima S, Boonchird C. Characterization and gene expression profiles of thermotolerant Saccharomyces cerevisiae isolates from Thai fruits. J Biosci Bioeng. 2012;114(2):144–149. doi: 10.1016/j.jbiosc.2012.03.012. - DOI - PubMed
-
- Caputi A, Ueda M, Brown T. Spectrophotometric determination of ethanol in wine. Am J Enol Vitic. 1968;19(3):160–165. doi: 10.5344/ajev.1968.19.3.160. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
