Heterologous versus homologous boosting elicits qualitatively distinct, BA.5-cross-reactive T cells in transplant recipients
- PMID: 37104041
- PMCID: PMC10322695
- DOI: 10.1172/jci.insight.168470
Heterologous versus homologous boosting elicits qualitatively distinct, BA.5-cross-reactive T cells in transplant recipients
Abstract
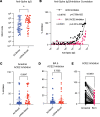
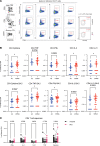
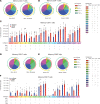
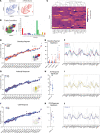
BackgroundThe SARS-CoV-2 Omicron BA.5 subvariant escapes vaccination-induced neutralizing antibodies because of mutations in the spike (S) protein. Solid organ transplant recipients (SOTRs) develop high COVID-19 morbidity and poor Omicron variant recognition after COVID-19 vaccination. T cell responses may provide a second line of defense. Therefore, understanding which vaccine regimens induce robust, conserved T cell responses is critical.MethodsWe evaluated anti-S IgG titers, subvariant pseudo-neutralization, and S-specific CD4+ and CD8+ T cell responses from SOTRs in a national, prospective, observational trial (n = 75). Participants were selected if they received 3 doses of mRNA (homologous boosting) or 2 doses of mRNA followed by Ad26.COV2.S (heterologous boosting).ResultsHomologous boosting with 3 mRNA doses induced the highest anti-S IgG titers. However, antibodies induced by both vaccine regimens demonstrated lower pseudo-neutralization against BA.5 compared with the ancestral strain. In contrast, vaccine-induced S-specific T cells maintained cross-reactivity against BA.5 compared with ancestral recognition. Homologous boosting induced higher frequencies of activated polyfunctional CD4+ T cell responses, with polyfunctional IL-21+ peripheral T follicular helper cells increased in mRNA-1273 compared with BNT162b2. IL-21+ cells correlated with antibody titers. Heterologous boosting with Ad26.COV2.S did not increase CD8+ responses compared to homologous boosting.ConclusionBoosting with the ancestral strain can induce cross-reactive T cell responses against emerging variants in SOTRs, but alternative vaccine strategies are required to induce robust CD8+ T cell responses.FundingBen-Dov Family; NIH National Institute of Allergy and Infectious Diseases (NIAID) K24AI144954, NIAID K08AI156021, NIAID K23AI157893, NIAID U01AI138897, National Institute of Diabetes and Digestive and Kidney Diseases T32DK007713, and National Cancer Institute 1U54CA260492; Johns Hopkins Vice Dean of Research Support for COVID-19 Research in Immunopathogenesis; and Emory COVID-19 research repository.
Keywords: Adaptive immunity; COVID-19; Organ transplantation; T cells; Vaccines.
Conflict of interest statement
Figures



References
-
- Haidar G, et al. Prospective evaluation of Coronavirus Disease 2019 (COVID-19) vaccine responses across a broad spectrum of immunocompromising conditions: the COVID-19 vaccination in the immunocompromised study (COVICS) Clin Infect Dis. 2022;ciac103(1):e630–e644. doi: 10.1093/cid/ciac103. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

