C5aR1 signaling triggers lung immunopathology in COVID-19 through neutrophil extracellular traps
- PMID: 37104043
- PMCID: PMC10266787
- DOI: 10.1172/JCI163105
C5aR1 signaling triggers lung immunopathology in COVID-19 through neutrophil extracellular traps
Abstract
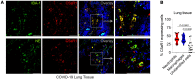
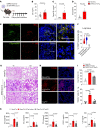
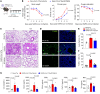
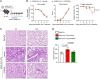
Patients with severe COVID-19 develop acute respiratory distress syndrome (ARDS) that may progress to cytokine storm syndrome, organ dysfunction, and death. Considering that complement component 5a (C5a), through its cellular receptor C5aR1, has potent proinflammatory actions and plays immunopathological roles in inflammatory diseases, we investigated whether the C5a/C5aR1 pathway could be involved in COVID-19 pathophysiology. C5a/C5aR1 signaling increased locally in the lung, especially in neutrophils of critically ill patients with COVID-19 compared with patients with influenza infection, as well as in the lung tissue of K18-hACE2 Tg mice (Tg mice) infected with SARS-CoV-2. Genetic and pharmacological inhibition of C5aR1 signaling ameliorated lung immunopathology in Tg-infected mice. Mechanistically, we found that C5aR1 signaling drives neutrophil extracellular traps-dependent (NETs-dependent) immunopathology. These data confirm the immunopathological role of C5a/C5aR1 signaling in COVID-19 and indicate that antagonists of C5aR1 could be useful for COVID-19 treatment.
Keywords: COVID-19; Complement; Inflammation; Innate immunity; Molecular pathology.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

