Enhanced evasion of neutralizing antibody response by Omicron XBB.1.5, CH.1.1, and CA.3.1 variants
- PMID: 37104089
- PMCID: PMC10279473
- DOI: 10.1016/j.celrep.2023.112443
Enhanced evasion of neutralizing antibody response by Omicron XBB.1.5, CH.1.1, and CA.3.1 variants
Abstract
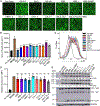
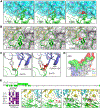
Omicron subvariants continuingly challenge current vaccination strategies. Here, we demonstrate nearly complete escape of the XBB.1.5, CH.1.1, and CA.3.1 variants from neutralizing antibodies stimulated by three doses of mRNA vaccine or by BA.4/5 wave infection, but neutralization is rescued by a BA.5-containing bivalent booster. CH.1.1 and CA.3.1 show strong immune escape from monoclonal antibody S309. Additionally, XBB.1.5, CH.1.1, and CA.3.1 spike proteins exhibit increased fusogenicity and enhanced processing compared with BA.2. Homology modeling reveals the key roles of G252V and F486P in the neutralization resistance of XBB.1.5, with F486P also enhancing receptor binding. Further, K444T/M and L452R in CH.1.1 and CA.3.1 likely drive escape from class II neutralizing antibodies, whereas R346T and G339H mutations could confer the strong neutralization resistance of these two subvariants to S309-like antibodies. Overall, our results support the need for administration of the bivalent mRNA vaccine and continued surveillance of Omicron subvariants.
Keywords: CA.3.1; CH.1.1; CP: Immunology; CP: Microbiology; Omicron subvariants; XBB.1.5; fusogenicity; immune evasion; mAb S309; neutralizing antibody.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Update of
-
Extraordinary Evasion of Neutralizing Antibody Response by Omicron XBB.1.5, CH.1.1 and CA.3.1 Variants.bioRxiv [Preprint]. 2023 Jan 17:2023.01.16.524244. doi: 10.1101/2023.01.16.524244. bioRxiv. 2023. Update in: Cell Rep. 2023 May 30;42(5):112443. doi: 10.1016/j.celrep.2023.112443. PMID: 36711991 Free PMC article. Updated. Preprint.
References
-
- Yuan S, Ye Z-W, Liang R, Tang K, Zhang AJ, Lu G, Ong CP, Man Poon VK, Chan CCS, Mok BWY, et al. (2022). Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters.Science 377, 428–433. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

