Toward Practical Integration of Omic and Imaging Data in Co-Clinical Trials
- PMID: 37104137
- PMCID: PMC10144684
- DOI: 10.3390/tomography9020066
Toward Practical Integration of Omic and Imaging Data in Co-Clinical Trials
Abstract
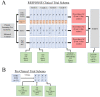
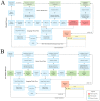
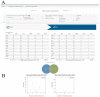
Co-clinical trials are the concurrent or sequential evaluation of therapeutics in both patients clinically and patient-derived xenografts (PDX) pre-clinically, in a manner designed to match the pharmacokinetics and pharmacodynamics of the agent(s) used. The primary goal is to determine the degree to which PDX cohort responses recapitulate patient cohort responses at the phenotypic and molecular levels, such that pre-clinical and clinical trials can inform one another. A major issue is how to manage, integrate, and analyze the abundance of data generated across both spatial and temporal scales, as well as across species. To address this issue, we are developing MIRACCL (molecular and imaging response analysis of co-clinical trials), a web-based analytical tool. For prototyping, we simulated data for a co-clinical trial in "triple-negative" breast cancer (TNBC) by pairing pre- (T0) and on-treatment (T1) magnetic resonance imaging (MRI) from the I-SPY2 trial, as well as PDX-based T0 and T1 MRI. Baseline (T0) and on-treatment (T1) RNA expression data were also simulated for TNBC and PDX. Image features derived from both datasets were cross-referenced to omic data to evaluate MIRACCL functionality for correlating and displaying MRI-based changes in tumor size, vascularity, and cellularity with changes in mRNA expression as a function of treatment.
Keywords: breast cancer; cancer informatics; cancer modeling; magnetic resonance imaging (MRI); multi-omics; radiomics.
Conflict of interest statement
M.T.L. is founder of, and an uncompensated limited partner in, StemMed Ltd. and a founder of, and uncompensated manager in StemMed Holdings, its general partner. M.T.L. is also a founder of, and equity holder in, Tvardi Therapeutics Inc. L.E.D. is a compensated employee of StemMed Ltd. Some PDXs are exclusively licensed to StemMed Ltd., resulting in royalty income to L.E.D.
Figures



References
-
- Petrosyan V., Dobrolecki L.E., Thistlethwaite L., Lewis A.N., Sallas C., Srinivasan R.R., Lei J.T., Kovacevic V., Obradovic P., Ellis M.J., et al. Identifying biomarkers of differential chemotherapy response in TNBC patient-Derived xenografts with a CTD/WGCNA approach. iScience. 2023;26:105799. doi: 10.1016/j.isci.2022.105799. - DOI - PMC - PubMed
-
- Shoghi K.I., Badea C.T., Blocker S.J., Chenevert T.L., Laforest R., Lewis M.T., Luker G.D., Manning H.C., Marcus D.S., Mowery Y.M., et al. Co-Clinical imaging resource program (CIRP): Bridging the translational divide to advance precision medicine. Tomography. 2020;6:273–287. doi: 10.18383/j.tom.2020.00023. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

