Analysis of the Effect of Plutella xylostella Polycalin and ABCC2 Transporter on Cry1Ac Susceptibility by CRISPR/Cas9-Mediated Knockout
- PMID: 37104211
- PMCID: PMC10145054
- DOI: 10.3390/toxins15040273
Analysis of the Effect of Plutella xylostella Polycalin and ABCC2 Transporter on Cry1Ac Susceptibility by CRISPR/Cas9-Mediated Knockout
Abstract
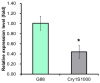
Many insects, including the Plutella xylostella (L.), have developed varying degrees of resistance to many insecticides, including Bacillus thuringiensis (Bt) toxins, the bioinsecticides derived from Bt. The polycalin protein is one of the potential receptors for Bt toxins, and previous studies have confirmed that the Cry1Ac toxin can bind to the polycalin protein of P. xylostella, but whether polycalin is associated with the resistance of Bt toxins remains controversial. In this study, we compared the midgut of larvae from Cry1Ac-susceptible and -resistant strains, and found that the expression of the Pxpolycalin gene was largely reduced in the midgut of the resistant strains. Moreover, the spatial and temporal expression patterns of Pxpolycalin showed that it was mainly expressed in the larval stage and midgut tissue. However, genetic linkage experiments showed that the Pxpolycalin gene and its transcript level were not linked to Cry1Ac resistance, whereas both the PxABCC2 gene and its transcript levels were linked to Cry1Ac resistance. The larvae fed on a diet containing the Cry1Ac toxin showed no significant change in the expression of the Pxpolycalin gene in a short term. Furthermore, the knockout of polycalin and ATP-binding cassette transporter subfamily C2 (ABCC2) genes separately by CRISPR/Cas9 technology resulted in resistance to decreased susceptibility to Cry1Ac toxin. Our results provide new insights into the potential role of polycalin and ABCC2 proteins in Cry1Ac resistance and the mechanism underlying the resistance of insects to Bt toxins.
Keywords: ABCC2; Bacillus thuringiensis; CRISPR/Cas9; polycalin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
CRISPR/Cas9-mediated knockout of both the PxABCC2 and PxABCC3 genes confers high-level resistance to Bacillus thuringiensis Cry1Ac toxin in the diamondback moth, Plutella xylostella (L.).Insect Biochem Mol Biol. 2019 Apr;107:31-38. doi: 10.1016/j.ibmb.2019.01.009. Epub 2019 Jan 30. Insect Biochem Mol Biol. 2019. PMID: 30710623
-
Downregulation of APN1 and ABCC2 mutation in Bt Cry1Ac-resistant Trichoplusia ni are genetically independent.Appl Environ Microbiol. 2024 Oct 23;90(10):e0074224. doi: 10.1128/aem.00742-24. Epub 2024 Sep 18. Appl Environ Microbiol. 2024. PMID: 39291983 Free PMC article.
-
Synergism of Bacillus thuringiensis Toxin Cry1Ac by a Fragment of Toxin-Binding Polycalin from Plutella xylostella.J Agric Food Chem. 2021 Oct 13;69(40):11816-11824. doi: 10.1021/acs.jafc.1c03156. Epub 2021 Oct 1. J Agric Food Chem. 2021. PMID: 34596393
-
[Advances in receptor-mediated resistance mechanisms of Lepidopteran insects to Bacillus thuringiensis toxin].Sheng Wu Gong Cheng Xue Bao. 2022 May 25;38(5):1809-1823. doi: 10.13345/j.cjb.210834. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 35611730 Review. Chinese.
-
Utilization of Diverse Molecules as Receptors by Cry Toxin and the Promiscuous Nature of Receptor-Binding Sites Which Accounts for the Diversity.Biomolecules. 2024 Apr 1;14(4):425. doi: 10.3390/biom14040425. Biomolecules. 2024. PMID: 38672442 Free PMC article. Review.
Cited by
-
A zinc finger protein shapes the temperature adaptability of a cosmopolitan pest.Open Biol. 2025 Apr;15(4):240346. doi: 10.1098/rsob.240346. Epub 2025 Apr 9. Open Biol. 2025. PMID: 40199340 Free PMC article.
-
CRISPR/Cas9 Genome Editing in the Diamondback Moth: Current Progress, Challenges, and Prospects.Int J Mol Sci. 2025 Feb 11;26(4):1515. doi: 10.3390/ijms26041515. Int J Mol Sci. 2025. PMID: 40003981 Free PMC article. Review.
References
-
- Usta C. In: Microorganisms in Biological Pest Control—A Review (Bacterial Toxin Application and Effect of Environmental Factors) Silva-Opps M., editor. InTech; Rijeka, Croatia: 1976. pp. 515–518.
-
- Tabashnik B.E., Cushing N.L., Finson N., Johnson M.W. Field development of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae) J. Econ. Entomol. 1990;83:1671–1676. doi: 10.1093/jee/83.5.1671. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

