RAD51 bypasses the CMG helicase to promote replication fork reversal
- PMID: 37104614
- PMCID: PMC10302453
- DOI: 10.1126/science.add7328
RAD51 bypasses the CMG helicase to promote replication fork reversal
Abstract
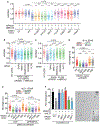
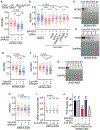
Replication fork reversal safeguards genome integrity as a replication stress response. DNA translocases and the RAD51 recombinase catalyze reversal. However, it remains unknown why RAD51 is required and what happens to the replication machinery during reversal. We find that RAD51 uses its strand exchange activity to circumvent the replicative helicase, which remains bound to the stalled fork. RAD51 is not required for fork reversal if the helicase is unloaded. Thus, we propose that RAD51 creates a parental DNA duplex behind the helicase that is used as a substrate by the DNA translocases for branch migration to create a reversed fork structure. Our data explain how fork reversal happens while maintaining the helicase in a position poised to restart DNA synthesis and complete genome duplication.
Conflict of interest statement
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

