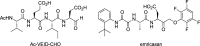Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6
- PMID: 37104712
- PMCID: PMC10176470
- DOI: 10.1021/jacs.2c12240
Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6
Abstract
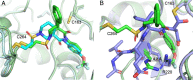
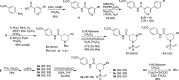
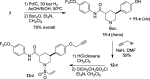
Caspases are a family of cysteine-dependent proteases with important cellular functions in inflammation and apoptosis, while also implicated in human diseases. Classical chemical tools to study caspase functions lack selectivity for specific caspase family members due to highly conserved active sites and catalytic machinery. To overcome this limitation, we targeted a non-catalytic cysteine residue (C264) unique to caspase-6 (C6), an enigmatic and understudied caspase isoform. Starting from disulfide ligands identified in a cysteine trapping screen, we used a structure-informed covalent ligand design to produce potent, irreversible inhibitors (3a) and chemoproteomic probes (13-t) of C6 that exhibit unprecedented selectivity over other caspase family members and high proteome selectivity. This approach and the new tools described will enable rigorous interrogation of the role of caspase-6 in developmental biology and in inflammatory and neurodegenerative diseases.
Conflict of interest statement
The authors declare the following competing financial interest(s): M.R.A. and A.R.R. are cofounders and advisors to Elgia Therapeutics.
Figures