Expression-based subtypes define pathologic response to neoadjuvant immune-checkpoint inhibitors in muscle-invasive bladder cancer
- PMID: 37105962
- PMCID: PMC10140274
- DOI: 10.1038/s41467-023-37568-9
Expression-based subtypes define pathologic response to neoadjuvant immune-checkpoint inhibitors in muscle-invasive bladder cancer
Abstract
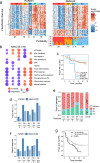
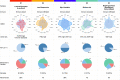
Checkpoint immunotherapy (CPI) has increased survival for some patients with advanced-stage bladder cancer (BCa). However, most patients do not respond. Here, we characterized the tumor and immune microenvironment in pre- and post-treatment tumors from the PURE01 neoadjuvant pembrolizumab immunotherapy trial, using a consolidative approach that combined transcriptional and genetic profiling with digital spatial profiling. We identify five distinctive genetic and transcriptomic programs and validate these in an independent neoadjuvant CPI trial to identify the features of response or resistance to CPI. By modeling the regulatory network, we identify the histone demethylase KDM5B as a repressor of tumor immune signaling pathways in one resistant subtype (S1, Luminal-excluded) and demonstrate that inhibition of KDM5B enhances immunogenicity in FGFR3-mutated BCa cells. Our study identifies signatures associated with response to CPI that can be used to molecularly stratify patients and suggests therapeutic alternatives for subtypes with poor response to neoadjuvant immunotherapy.
© 2023. The Author(s).
Conflict of interest statement
J.J.M. participated in advisory boards for AstraZeneca, Astellas/Seagen, BMS, Janssen, Prokarium, Pfizer, Merck, and UroGen. A.N. is a consultant for Merck, Astra Zeneca, Janssen, Incyte, Roche, Rainier Therapeutics, Clovis Oncology, Bayer, Astellas/Seattle Genetics, Ferring, and Immunomedics. Travel expenses/Honoraria: Roche, Merck, Astra Zeneca, and Janssen. L.M.: Speaker compensation: Merck; Travel expenses and accommodation: Janssen; Research funding: AstraZeneca. AdR is a member of the scientific advisory board of Qlucore and cofounder of Minos Biosciences. A.G.R., K.M., L.C., K.M., L.A., Y.Y., M.C., CG., V.N., V.T., B.C., F.M., and T.P. have no disclosures.
Figures





Comment in
-
Uro-Science.J Urol. 2024 Jan;211(1):194-195. doi: 10.1097/JU.0000000000003710. Epub 2023 Oct 20. J Urol. 2024. PMID: 37861082 No abstract available.
References
-
- Rosenberg JE, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

