DNA polymerase POLD1 promotes proliferation and metastasis of bladder cancer by stabilizing MYC
- PMID: 37105989
- PMCID: PMC10140023
- DOI: 10.1038/s41467-023-38160-x
DNA polymerase POLD1 promotes proliferation and metastasis of bladder cancer by stabilizing MYC
Abstract
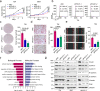
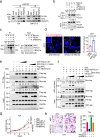
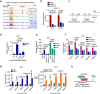
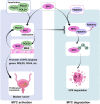
To date, most studies on the DNA polymerase, POLD1, have focused on the effect of POLD1 inactivation mutations in tumors. However, the implications of high POLD1 expression in tumorigenesis remains elusive. Here, we determine that POLD1 has a pro-carcinogenic role in bladder cancer (BLCA) and is associated to the malignancy and prognosis of BLCA. Our studies demonstrate that POLD1 promotes the proliferation and metastasis of BLCA via MYC. Mechanistically, POLD1 stabilizes MYC in a manner independent of its' DNA polymerase activity. Instead, POLD1 attenuates FBXW7-mediated ubiquitination degradation of MYC by directly binding to the MYC homology box 1 domain competitively with FBXW7. Moreover, we find that POLD1 forms a complex with MYC to promote the transcriptional activity of MYC. In turn, MYC increases expression of POLD1, forming a POLD1-MYC positive feedback loop to enhance the pro-carcinogenic effect of POLD1-MYC on BLCA. Overall, our study identifies POLD1 as a promotor of BCLA via a MYC driven mechanism and suggest its potential as biomarker for BLCA.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. 2021;71:209–249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

