Single duplex DNA sequencing with CODEC detects mutations with high sensitivity
- PMID: 37106072
- PMCID: PMC10181940
- DOI: 10.1038/s41588-023-01376-0
Single duplex DNA sequencing with CODEC detects mutations with high sensitivity
Abstract
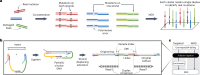
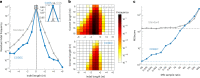
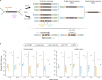
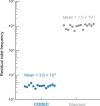
Detecting mutations from single DNA molecules is crucial in many fields but challenging. Next-generation sequencing (NGS) affords tremendous throughput but cannot directly sequence double-stranded DNA molecules ('single duplexes') to discern the true mutations on both strands. Here we present Concatenating Original Duplex for Error Correction (CODEC), which confers single duplex resolution to NGS. CODEC affords 1,000-fold higher accuracy than NGS, using up to 100-fold fewer reads than duplex sequencing. CODEC revealed mutation frequencies of 2.72 × 10-8 in sperm of a 39-year-old individual, and somatic mutations acquired with age in blood cells. CODEC detected genome-wide, clonal hematopoiesis mutations from single DNA molecules, single mutated duplexes from tumor genomes and liquid biopsies, microsatellite instability with 10-fold greater sensitivity and mutational signatures, and specific tumor mutations with up to 100-fold fewer reads. CODEC enables more precise genetic testing and reveals biologically significant mutations, which are commonly obscured by NGS errors.
© 2023. The Author(s).
Conflict of interest statement
V.A.A., J.H.B., R.L. and G.M.M. have filed a patent application (РСТ/US2021/062966) on this method. T.R.G. has paid scientific advisory roles and equity in Dewpoint Therapeutics and Anji Pharmaceuticals, holds founder’s equity in Sherlock Biosciences, is a paid advisor to Braidwell Inc. and has research funding from Bayer HealthCare, Calico Life Sciences and Novo Holdings. J.E.S. is the key opinion leader for ForTec Medical. The remaining authors declare no competing interests.
Figures










References
-
- Beaubier N, et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat. Biotechnol. 2019;37:1351–1360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

