PlaASDB: a comprehensive database of plant alternative splicing events in response to stress
- PMID: 37106367
- PMCID: PMC10134664
- DOI: 10.1186/s12870-023-04234-7
PlaASDB: a comprehensive database of plant alternative splicing events in response to stress
Abstract
Background: Alternative splicing (AS) is a co-transcriptional regulatory mechanism of plants in response to environmental stress. However, the role of AS in biotic and abiotic stress responses remains largely unknown. To speed up our understanding of plant AS patterns under different stress responses, development of informative and comprehensive plant AS databases is highly demanded.
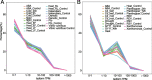
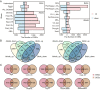
Description: In this study, we first collected 3,255 RNA-seq data under biotic and abiotic stresses from two important model plants (Arabidopsis and rice). Then, we conducted AS event detection and gene expression analysis, and established a user-friendly plant AS database termed PlaASDB. By using representative samples from this highly integrated database resource, we compared AS patterns between Arabidopsis and rice under abiotic and biotic stresses, and further investigated the corresponding difference between AS and gene expression. Specifically, we found that differentially spliced genes (DSGs) and differentially expressed genes (DEG) share very limited overlapping under all kinds of stresses, suggesting that gene expression regulation and AS seemed to play independent roles in response to stresses. Compared with gene expression, Arabidopsis and rice were more inclined to have conserved AS patterns under stress conditions.
Conclusion: PlaASDB is a comprehensive plant-specific AS database that mainly integrates the AS and gene expression data of Arabidopsis and rice in stress response. Through large-scale comparative analyses, the global landscape of AS events in Arabidopsis and rice was observed. We believe that PlaASDB could help researchers understand the regulatory mechanisms of AS in plants under stresses more conveniently. PlaASDB is freely accessible at http://zzdlab.com/PlaASDB/ASDB/index.html .
Keywords: Alternative splicing; Arabidopsis; Plant; RNA-Seq data; Rice; Stress response.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

