Proteomic Profiling of Fallopian Tube-Derived Extracellular Vesicles Using a Microfluidic Tissue-on-Chip System
- PMID: 37106610
- PMCID: PMC10135590
- DOI: 10.3390/bioengineering10040423
Proteomic Profiling of Fallopian Tube-Derived Extracellular Vesicles Using a Microfluidic Tissue-on-Chip System
Abstract
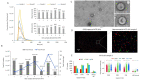
The human fallopian tube epithelium (hFTE) is the site of fertilization, early embryo development, and the origin of most high-grade serous ovarian cancers (HGSOCs). Little is known about the content and functions of hFTE-derived small extracellular vesicles (sEVs) due to the limitations of biomaterials and proper culture methods. We have established a microfluidic platform to culture hFTE for EV collection with adequate yield for mass spectrometry-based proteomic profiling, and reported 295 common hFTE sEV proteins for the first time. These proteins are associated with exocytosis, neutrophil degranulation, and wound healing, and some are crucial for fertilization processes. In addition, by correlating sEV protein profiles with hFTE tissue transcripts characterized using GeoMx® Cancer Transcriptome Atlas, spatial transcriptomics analysis revealed cell-type-specific transcripts of hFTE that encode sEVs proteins, among which, FLNA, TUBB, JUP, and FLNC were differentially expressed in secretory cells, the precursor cells for HGSOC. Our study provides insights into the establishment of the baseline proteomic profile of sEVs derived from hFTE tissue, and its correlation with hFTE lineage-specific transcripts, which can be used to evaluate whether the fallopian tube shifts its sEV cargo during ovarian cancer carcinogenesis and the role of sEV proteins in fallopian tube reproductive functions.
Keywords: digital spatial imaging; extracellular vesicles; fallopian tube; microfluidic culture; proteomics.
Conflict of interest statement
A.K.G. is a co-founder of Sinochips Diagnostics, serves as a scientific advisory board member to Biovica, Clara Biotech, and Sinochips Diagnostics, and receives research funding from Predicine, Inc. and VITRAC Therapeutics. The other authors report no conflicts of interest.
Figures




References
-
- Almiñana C., Corbin E., Tsikis G., Alcântara-Neto A.S., Labas V., Reynaud K., Galio L., Uzbekov R., Garanina A.S., Druart X., et al. Oviduct Extracellular Vesicles Protein Content and Their Role during Oviduct–Embryo Cross-Talk. Reproduction. 2017;154:253–268. doi: 10.1530/REP-17-0054. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

