CalcAMP: A New Machine Learning Model for the Accurate Prediction of Antimicrobial Activity of Peptides
- PMID: 37107088
- PMCID: PMC10135148
- DOI: 10.3390/antibiotics12040725
CalcAMP: A New Machine Learning Model for the Accurate Prediction of Antimicrobial Activity of Peptides
Abstract
To combat infection by microorganisms host organisms possess a primary arsenal via the innate immune system. Among them are defense peptides with the ability to target a wide range of pathogenic organisms, including bacteria, viruses, parasites, and fungi. Here, we present the development of a novel machine learning model capable of predicting the activity of antimicrobial peptides (AMPs), CalcAMP. AMPs, in particular short ones (<35 amino acids), can become an effective solution to face the multi-drug resistance issue arising worldwide. Whereas finding potent AMPs through classical wet-lab techniques is still a long and expensive process, a machine learning model can be useful to help researchers to rapidly identify whether peptides present potential or not. Our prediction model is based on a new data set constructed from the available public data on AMPs and experimental antimicrobial activities. CalcAMP can predict activity against both Gram-positive and Gram-negative bacteria. Different features either concerning general physicochemical properties or sequence composition have been assessed to retrieve higher prediction accuracy. CalcAMP can be used as an promising prediction asset to identify short AMPs among given peptide sequences.
Keywords: antimicrobial peptides; antimicrobial resistance; artificial intelligence; bacteria; drug discovery; machine learning.
Conflict of interest statement
Madam Therapeutics is a commercial company that aims to put these type of AMPs on the market.
Figures

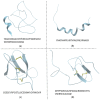
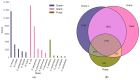

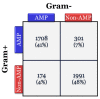
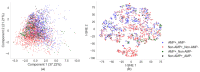




References
-
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. - DOI - PubMed
-
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. [(accessed on 9 November 2022)]. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-....
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

