By-Product Extracts from Castanea sativa Counteract Hallmarks of Neuroinflammation in a Microglial Model
- PMID: 37107183
- PMCID: PMC10135167
- DOI: 10.3390/antiox12040808
By-Product Extracts from Castanea sativa Counteract Hallmarks of Neuroinflammation in a Microglial Model
Abstract
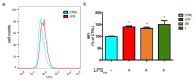
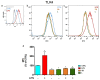
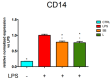
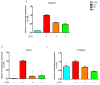
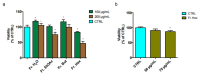
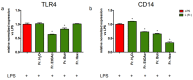
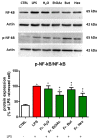
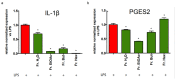
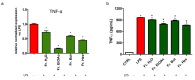
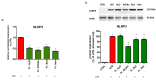
Castanea sativa is very common in Italy, and the large amount of waste material generated during chestnut processing has a high environmental impact. Several studies demonstrated that chestnut by-products are a good source of bioactive compounds, mainly endowed with antioxidant properties. This study further investigates the anti-neuroinflammatory effect of chestnut leaf and spiny bur extracts, together with the deepest phytochemical characterisation (by NMR and MS) of active biomolecules contained in leaf extracts, which resulted in being more effective than spiny bur ones. BV-2 microglial cells stimulated with lipopolysaccharide (LPS) were used as a model of neuroinflammation. In BV-2 cells pre-treated with chestnut extracts, LPS signalling is partially blocked via the reduced expression of TLR4 and CD14 as well as the expression of LPS-induced inflammatory markers. Leaf extract fractions revealed the presence of specific flavonoids, such as isorhamnetin glucoside, astragalin, myricitrin, kaempferol 3-rhamnosyl (1-6)(2″-trans-p-coumaroyl)hexoside, tiliroside and unsaturated fatty acids, all of which could be responsible for the observed anti-neuroinflammatory effects. Interestingly, the kaempferol derivative has been identified in chestnut for the first time. In conclusion, the exploitation of chestnut by-products is suitable for the achievement of two goals: satisfaction of consumers' demand for new, natural bio-active compounds and valorisation of by-products.
Keywords: BV-2; Castanea sativa; NF-kB; Toll-like receptor 4; chestnut by-products; flavonoids; microglia; neuroinflammation; waste valorisation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Barreira J., Ferreira I., Oliveira M., Pereira J. Antioxidant Activities of the Extracts from Chestnut Flower, Leaf, Skins and Fruit. Food Chem. 2008;107:1106–1113. doi: 10.1016/j.foodchem.2007.09.030. - DOI
-
- Chiarini A., Micucci M., Malaguti M., Budriesi R., Ioan P., Lenzi M., Fimognari C., Gallina Toschi T., Comandini P., Hrelia S. Sweet Chestnut (Castanea sativa Mill.) Bark Extract: Cardiovascular Activity and Myocyte Protection against Oxidative Damage. Oxidative Med. Cell. Longev. 2013;2013:471790. doi: 10.1155/2013/471790. - DOI - PMC - PubMed
-
- Squillaci G., Apone F., Sena L.M., Carola A., Tito A., Bimonte M., Lucia A.D., Colucci G., Cara F.L., Morana A. Chestnut (Castanea sativa Mill.) Industrial Wastes as a Valued Bioresource for the Production of Active Ingredients. Process Biochem. 2018;64:228–236. doi: 10.1016/j.procbio.2017.09.017. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

