Ovule Transcriptome Analysis Discloses Deregulation of Genes and Pathways in Sexual and Apomictic Limonium Species (Plumbaginaceae)
- PMID: 37107659
- PMCID: PMC10137852
- DOI: 10.3390/genes14040901
Ovule Transcriptome Analysis Discloses Deregulation of Genes and Pathways in Sexual and Apomictic Limonium Species (Plumbaginaceae)
Abstract
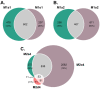
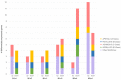
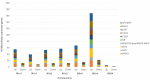
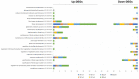
The genus Limonium Mill. (sea lavenders) includes species with sexual and apomixis reproductive strategies, although the genes involved in these processes are unknown. To explore the mechanisms beyond these reproduction modes, transcriptome profiling of sexual, male sterile, and facultative apomictic species was carried out using ovules from different developmental stages. In total, 15,166 unigenes were found to be differentially expressed with apomictic vs. sexual reproduction, of which 4275 were uniquely annotated using an Arabidopsis thaliana database, with different regulations according to each stage and/or species compared. Gene ontology (GO) enrichment analysis indicated that genes related to tubulin, actin, the ubiquitin degradation process, reactive oxygen species scavenging, hormone signaling such as the ethylene signaling pathway and gibberellic acid-dependent signal, and transcription factors were found among differentially expressed genes (DEGs) between apomictic and sexual plants. We found that 24% of uniquely annotated DEGs were likely to be implicated in flower development, male sterility, pollen formation, pollen-stigma interactions, and pollen tube formation. The present study identifies candidate genes that are highly associated with distinct reproductive modes and sheds light on the molecular mechanisms of apomixis expression in Limonium sp.
Keywords: Illumina sequencing; MADS-box; apomixis; floral development; functional annotation; male sterility; sea lavenders; sexual reproduction; transcription factors.
Conflict of interest statement
The authors declare no conflict of interest. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Figures

References
-
- Asker S.E., Jerling L. Apomixis in Plants. 1st ed. CRC Press; Boca Raton, FL, USA: 1992.
-
- Richards A.J. Plant Breeding Systems. Chapman and Hall; London, UK: 1997.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

