Comparative Transcriptomics in Atlantic Salmon Head Kidney and SHK-1 Cell Line Exposed to the Sea Louse Cr-Cathepsin
- PMID: 37107663
- PMCID: PMC10138087
- DOI: 10.3390/genes14040905
Comparative Transcriptomics in Atlantic Salmon Head Kidney and SHK-1 Cell Line Exposed to the Sea Louse Cr-Cathepsin
Abstract
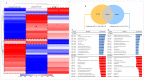
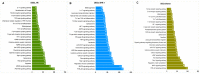
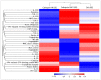
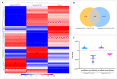
The development of vaccines against sea lice in salmon farming is complex, expensive, and takes several years for commercial availability. Recently, transcriptome studies in sea louse have provided valuable information for identifying relevant molecules with potential use for fish vaccines. However, the bottleneck is the in vivo testing of recombinant protein candidates, the dosage, and the polyvalent formulation strategies. This study explored a cell-based approach to prospect antigens as candidate vaccines against sea lice by comparison with immunized fish. Herein, SHK-1 cells and Atlantic salmon head kidney tissue were exposed to the antigen cathepsin identified from the sea louse Caligus rogercresseyi. The cathepsin protein was cloned and recombinantly expressed in Escherichia coli, and then SHK-1 cell lines were stimulated with 100 ng/mL cathepsin recombinant for 24 h. In addition, Atlantic salmons were vaccinated with 30 ug/mL recombinant protein, and head kidney samples were then collected 30 days post-immunization. SHK-1 cells and salmon head kidney exposed to cathepsin were analyzed by Illumina RNA sequencing. The statistical comparisons showed differences in the transcriptomic profiles between SHK-1 cells and the salmon head kidney. However, 24.15% of the differentially expressed genes were shared. Moreover, putative gene regulation through lncRNAs revealed tissue-specific transcription patterns. The top 50 up and downregulated lncRNAs were highly correlated with genes involved in immune response, iron homeostasis, pro-inflammatory cytokines, and apoptosis. Also, highly enriched pathways related to the immune system and signal transduction were shared between both tissues. These findings highlight a novel approach to evaluating candidate antigens for sea lice vaccine development, improving the antigens screening in the SHK-1 cell line model.
Keywords: C. rogercresseyi; immune response; recombinant protein; transcriptome response.
Conflict of interest statement
The authors declare that they have no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- Sneeringer S., Bowman M., Clancy M. The U.S. and EU Animal Pharmaceutical Industries in the Age of Antibiotic Resistance. USDA; Washington DC, USA: 2019.
-
- FAO . The State of World Fisheries and Aquaculture 2022. FAO; Rome, Italy: 2022.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

