Long-Term Transcriptomic Changes and Cardiomyocyte Hyperpolyploidy after Lactose Intolerance in Neonatal Rats
- PMID: 37108224
- PMCID: PMC10138443
- DOI: 10.3390/ijms24087063
Long-Term Transcriptomic Changes and Cardiomyocyte Hyperpolyploidy after Lactose Intolerance in Neonatal Rats
Abstract
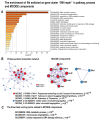
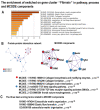
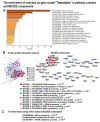
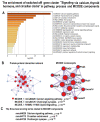
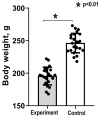
Many cardiovascular diseases originate from growth retardation, inflammation, and malnutrition during early postnatal development. The nature of this phenomenon is not completely understood. Here we aimed to verify the hypothesis that systemic inflammation triggered by neonatal lactose intolerance (NLI) may exert long-term pathologic effects on cardiac developmental programs and cardiomyocyte transcriptome regulation. Using the rat model of NLI triggered by lactase overloading with lactose and the methods of cytophotometry, image analysis, and mRNA-seq, we evaluated cardiomyocyte ploidy, signs of DNA damage, and NLI-associated long-term transcriptomic changes of genes and gene modules that differed qualitatively (i.e., were switched on or switched off) in the experiment vs. the control. Our data indicated that NLI triggers the long-term animal growth retardation, cardiomyocyte hyperpolyploidy, and extensive transcriptomic rearrangements. Many of these rearrangements are known as manifestations of heart pathologies, including DNA and telomere instability, inflammation, fibrosis, and reactivation of fetal gene program. Moreover, bioinformatic analysis identified possible causes of these pathologic traits, including the impaired signaling via thyroid hormone, calcium, and glutathione. We also found transcriptomic manifestations of increased cardiomyocyte polyploidy, such as the induction of gene modules related to open chromatin, e.g., "negative regulation of chromosome organization", "transcription" and "ribosome biogenesis". These findings suggest that ploidy-related epigenetic alterations acquired in the neonatal period permanently rewire gene regulatory networks and alter cardiomyocyte transcriptome. Here we provided first evidence indicating that NLI can be an important trigger of developmental programming of adult cardiovascular disease. The obtained results can help to develop preventive strategies for reducing the NLI-associated adverse effects of inflammation on the developing cardiovascular system.
Keywords: DNA instability; cardiomyocyte; developmental programming of adult heart diseases; fibrosis; glutathione deficiency; inflammation; neonatal lactose intolerance; polyploidy; qualitative transcriptome analysis; thyroid deficiency.
Conflict of interest statement
The authors declare no potential conflict of interest. The authors confirm that the data supporting the findings of this study are available within the paper and its Supplementary Materials.
Figures






References
-
- Harikrishnan S., Jeemon P., Mini G.K., Thankappan K.R., Sylaja P. GBD 2017 Causes of Death Collaborators Global, Regional, and National Age-Sex-Specific Mortality for 282 Causes of Death in 195 Countries and Territories, 1980–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1736–1788. doi: 10.1016/s0140-6736(18)32203-7. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

