Lipid-Sensing Receptor FFAR4 Modulates Pulmonary Epithelial Homeostasis following Immunogenic Exposures Independently of the FFAR4 Ligand Docosahexaenoic Acid (DHA)
- PMID: 37108233
- PMCID: PMC10138935
- DOI: 10.3390/ijms24087072
Lipid-Sensing Receptor FFAR4 Modulates Pulmonary Epithelial Homeostasis following Immunogenic Exposures Independently of the FFAR4 Ligand Docosahexaenoic Acid (DHA)
Abstract
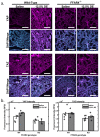
The role of pulmonary free fatty acid receptor 4 (FFAR4) is not fully elucidated and we aimed to clarify the impact of FFAR4 on the pulmonary immune response and return to homeostasis. We employed a known high-risk human pulmonary immunogenic exposure to extracts of dust from swine confinement facilities (DE). WT and Ffar4-null mice were repetitively exposed to DE via intranasal instillation and supplemented with docosahexaenoic acid (DHA) by oral gavage. We sought to understand if previous findings of DHA-mediated attenuation of the DE-induced inflammatory response are FFAR4-dependent. We identified that DHA mediates anti-inflammatory effects independent of FFAR4 expression, and that DE-exposed mice lacking FFAR4 had reduced immune cells in the airways, epithelial dysplasia, and impaired pulmonary barrier integrity. Analysis of transcripts using an immunology gene expression panel revealed a role for FFAR4 in lungs related to innate immune initiation of inflammation, cytoprotection, and immune cell migration. Ultimately, the presence of FFAR4 in the lung may regulate cell survival and repair following immune injury, suggestive of potential therapeutic directions for pulmonary disease.
Keywords: DHA; FFAR4; GPR120; exposure; inflammation; lung.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
DHA supplementation prevent the progression of NASH via GPR120 signaling.Eur J Pharmacol. 2018 Feb 5;820:31-38. doi: 10.1016/j.ejphar.2017.11.046. Epub 2017 Dec 6. Eur J Pharmacol. 2018. PMID: 29221950
-
Docosahexaenoic Acid Attenuates the Progression of Nonalcoholic Steatohepatitis by Suppressing the Adipocyte Inflammation via the G Protein-Coupled Receptor 120/Free Fatty Acid Receptor 4 Pathway.Pharmacology. 2022;107(5-6):330-338. doi: 10.1159/000522117. Epub 2022 Feb 21. Pharmacology. 2022. PMID: 35189618
-
Docosahexaenoic acid enhances amphiregulin-mediated bronchial epithelial cell repair processes following organic dust exposure.Am J Physiol Lung Cell Mol Physiol. 2018 Mar 1;314(3):L421-L431. doi: 10.1152/ajplung.00273.2017. Epub 2017 Nov 2. Am J Physiol Lung Cell Mol Physiol. 2018. PMID: 29097425 Free PMC article.
-
[Involvement of the Free Fatty Acid Receptor GPR120/FFAR4 in the Development of Nonalcoholic Steatohepatitis].Yakugaku Zasshi. 2019;139(9):1169-1175. doi: 10.1248/yakushi.19-00011-4. Yakugaku Zasshi. 2019. PMID: 31474633 Review. Japanese.
-
FFAR4: A New Player in Cardiometabolic Disease?Endocrinology. 2021 Aug 1;162(8):bqab111. doi: 10.1210/endocr/bqab111. Endocrinology. 2021. PMID: 34043793 Free PMC article. Review.
Cited by
-
Potential Therapeutic Exploitation of G Protein-Coupled Receptor 120 (GPR120/FFAR4) Signaling in Obesity-Related Metabolic Disorders.Int J Mol Sci. 2025 Mar 11;26(6):2501. doi: 10.3390/ijms26062501. Int J Mol Sci. 2025. PMID: 40141148 Free PMC article. Review.
References
-
- Nordgren T.M., Heires A.J., Bailey K.L., Katafiasz D.M., Toews M.L., Wichman C.S., Romberger D.J. Docosahexaenoic acid enhances amphiregulin-mediated bronchial epithelial cell repair processes following organic dust exposure. Am. J. Physiol. Lung Cell Mol. Physiol. 2018;314:L421–L431. doi: 10.1152/ajplung.00273.2017. - DOI - PMC - PubMed
-
- Skulas-Ray A.C., Wilson P.W.F., Harris W.S., Brinton E.A., Kris-Etherton P.M., Richter C.K., Jacobson T.A., Engler M.B., Miller M., Robinson J.G., et al. Omega-3 Fatty Acids for the Management of Hypertriglyceridemia: A Science Advisory From the American Heart Association. Circulation. 2019;140:e673–e691. doi: 10.1161/CIR.0000000000000709. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

