HSV-1 Glycoprotein D and Its Surface Receptors: Evaluation of Protein-Protein Interaction and Targeting by Triazole-Based Compounds through In Silico Approaches
- PMID: 37108255
- PMCID: PMC10138673
- DOI: 10.3390/ijms24087092
HSV-1 Glycoprotein D and Its Surface Receptors: Evaluation of Protein-Protein Interaction and Targeting by Triazole-Based Compounds through In Silico Approaches
Abstract
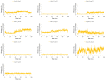
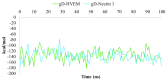
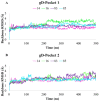
Protein-protein interactions (PPI) represent attractive targets for drug design. Thus, aiming at a deeper insight into the HSV-1 envelope glycoprotein D (gD), protein-protein docking and dynamic simulations of gD-HVEM and gD-Nectin-1 complexes were performed. The most stable complexes and the pivotal key residues useful for gD to anchor human receptors were identified and used as starting points for a structure-based virtual screening on a library of both synthetic and designed 1,2,3-triazole-based compounds. Their binding properties versus gD interface with HVEM and Nectin-1 along with their structure-activity relationships (SARs) were evaluated. Four [1,2,3]triazolo[4,5-b]pyridines were identified as potential HSV-1 gD inhibitors, for their good theoretical affinity towards all conformations of HSV-1 gD. Overall, this study suggests promising basis for the design of new antiviral agents targeting gD as a valuable strategy to prevent viral attachment and penetration into the host cell.
Keywords: 1,2,3-triazoles; HSV-1; docking; glycoprotein D; molecular dynamics simulations; protein–protein interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

