Assessing the Orthogonality of Phage-Encoded RNA Polymerases for Tailored Synthetic Biology Applications in Pseudomonas Species
- PMID: 37108338
- PMCID: PMC10138996
- DOI: 10.3390/ijms24087175
Assessing the Orthogonality of Phage-Encoded RNA Polymerases for Tailored Synthetic Biology Applications in Pseudomonas Species
Abstract
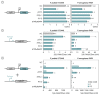
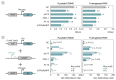
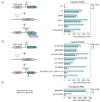
The phage T7 RNA polymerase (RNAP) and lysozyme form the basis of the widely used pET expression system for recombinant expression in the biotechnology field and as a tool in microbial synthetic biology. Attempts to transfer this genetic circuitry from Escherichia coli to non-model bacterial organisms with high potential have been restricted by the cytotoxicity of the T7 RNAP in the receiving hosts. We here explore the diversity of T7-like RNAPs mined directly from Pseudomonas phages for implementation in Pseudomonas species, thus relying on the co-evolution and natural adaptation of the system towards its host. By screening and characterizing different viral transcription machinery using a vector-based system in P. putida., we identified a set of four non-toxic phage RNAPs from phages phi15, PPPL-1, Pf-10, and 67PfluR64PP, showing a broad activity range and orthogonality to each other and the T7 RNAP. In addition, we confirmed the transcription start sites of their predicted promoters and improved the stringency of the phage RNAP expression systems by introducing and optimizing phage lysozymes for RNAP inhibition. This set of viral RNAPs expands the adaption of T7-inspired circuitry towards Pseudomonas species and highlights the potential of mining tailored genetic parts and tools from phages for their non-model host.
Keywords: Pseudomonas; RNA polymerase; T7-like phages; lysozyme; orthogonality.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Engineering a phi15-based expression system for stringent gene expression in Pseudomonas putida.Commun Biol. 2025 Feb 4;8(1):171. doi: 10.1038/s42003-025-07508-y. Commun Biol. 2025. PMID: 39905116 Free PMC article.
-
Host RNA polymerase inhibitors encoded by ϕKMV-like phages of Pseudomonas.Virology. 2013 Feb 5;436(1):67-74. doi: 10.1016/j.virol.2012.10.021. Epub 2012 Nov 3. Virology. 2013. PMID: 23127595
-
Transcriptomics-Driven Characterization of LUZ100, a T7-like Pseudomonas Phage with Temperate Features.mSystems. 2023 Apr 27;8(2):e0118922. doi: 10.1128/msystems.01189-22. Epub 2023 Feb 16. mSystems. 2023. PMID: 36794936 Free PMC article.
-
Multisubunit RNA Polymerases of Jumbo Bacteriophages.Viruses. 2020 Sep 23;12(10):1064. doi: 10.3390/v12101064. Viruses. 2020. PMID: 32977622 Free PMC article. Review.
-
Xenogeneic Regulation of the Bacterial Transcription Machinery.J Mol Biol. 2019 Sep 20;431(20):4078-4092. doi: 10.1016/j.jmb.2019.02.008. Epub 2019 Feb 15. J Mol Biol. 2019. PMID: 30776429 Review.
Cited by
-
Decoding and reengineering the promoter specificity of T7-like RNA polymerases based on phage genome sequences.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf140. doi: 10.1093/nar/gkaf140. Nucleic Acids Res. 2025. PMID: 40042813 Free PMC article.
-
Engineering a phi15-based expression system for stringent gene expression in Pseudomonas putida.Commun Biol. 2025 Feb 4;8(1):171. doi: 10.1038/s42003-025-07508-y. Commun Biol. 2025. PMID: 39905116 Free PMC article.
References
-
- Luo R., Lian G., Li H., Han H., Zhou D., Gong X. Ultrasensitive Sensing of T4 PNK Phosphatase Activity through Establishing a Novel Transcription-Based Signal Amplification Platform. Sens. Actuators B Chem. 2022;369:132269. doi: 10.1016/j.snb.2022.132269. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

