Identification and Functional Analysis of Tartrate-Resistant Acid Phosphatase Type 5b (TRAP5b) in Oreochromis niloticus
- PMID: 37108342
- PMCID: PMC10138680
- DOI: 10.3390/ijms24087179
Identification and Functional Analysis of Tartrate-Resistant Acid Phosphatase Type 5b (TRAP5b) in Oreochromis niloticus
Abstract
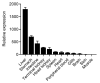
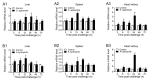
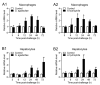
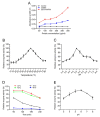
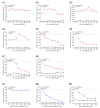
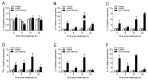
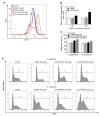
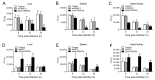
Tartrate-resistant acid phosphatase type 5 (TRAP5) is an enzyme that is highly expressed in activated macrophages and osteoclasts and plays important biological functions in mammalian immune defense systems. In the study, we investigated the functions of tartrate-resistant acid phosphatase type 5b from Oreochromis niloticus (OnTRAP5b). The OnTRAP5b gene has an open reading frame of 975 bp, which encodes a mature peptide consisting of 302 amino acids with a molecular weight of 33.448 kDa. The OnTRAP5b protein contains a metallophosphatase domain with metal binding and active sites. Phylogenetic analysis revealed that OnTRAP5b is clustered with TRAP5b of teleost fish and shares a high amino acid sequence similarity with other TRAP5b in teleost fish (61.73-98.15%). Tissues expression analysis showed that OnTRAP5b was most abundant in the liver and was also widely expressed in other tissues. Upon challenge with Streptococcus agalactiae and Aeromonas hydrophila in vivo and in vitro, the expression of OnTRAP5b was significantly up-regulated. Additionally, the purified recombinant OnTRAP5b ((r)OnTRAP5) protein exhibited optimal phosphatase activity at pH 5.0 and an ideal temperature of 50 °C. The Vmax, Km, and kcat of purified (r)OnTRAP5b were found to be 0.484 μmol × min-1 × mg-1, 2.112 mM, and 0.27 s-1 with respect to pNPP as a substrate, respectively. Its phosphatase activity was differentially affected by metal ions (K+, Na+, Mg2+, Ca2+, Mn2+, Cu2+, Zn2+, and Fe3+) and inhibitors (sodium tartrate, sodium fluoride, and EDTA). Furthermore, (r)OnTRAP5b was found to promote the expression of inflammatory-related genes in head kidney macrophages and induce reactive oxygen expression and phagocytosis. Moreover, OnTRAP5b overexpression and knockdown had a significant effect on bacterial proliferation in vivo. When taken together, our findings suggest that OnTRAP5b plays a significant role in the immune response against bacterial infection in Nile tilapia.
Keywords: Oreochromis niloticus; phagocytosis; phosphatase activity; reactive oxygen species; tartrate-resistant acid phosphatase type 5b (TRAP5b).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identification and characterization of a mannose-binding lectin from Nile tilapia (Oreochromis niloticus).Fish Shellfish Immunol. 2017 Aug;67:244-253. doi: 10.1016/j.fsi.2017.06.016. Epub 2017 Jun 7. Fish Shellfish Immunol. 2017. PMID: 28602737
-
Expression and functional characterization of collection-K1 from Nile tilapia (Oreochromis niloticus) in host innate immune defense.Mol Immunol. 2018 Nov;103:21-34. doi: 10.1016/j.molimm.2018.08.012. Epub 2018 Sep 3. Mol Immunol. 2018. PMID: 30189385
-
Expression and functional characterization of a mannose-binding lectin-associated serine protease-1 (MASP-1) from Nile tilapia (Oreochromis niloticus) in host defense against bacterial infection.Fish Shellfish Immunol. 2019 Aug;91:68-77. doi: 10.1016/j.fsi.2019.05.014. Epub 2019 May 13. Fish Shellfish Immunol. 2019. PMID: 31096060
-
LECT2 Protects Nile Tilapia (Oreochromis niloticus) Against Streptococcus agalatiae Infection.Front Immunol. 2021 May 20;12:667781. doi: 10.3389/fimmu.2021.667781. eCollection 2021. Front Immunol. 2021. PMID: 34093564 Free PMC article.
-
A C-type lectin (CL11X1-like) from Nile tilapia (Oreochromis niloticus) is involved in host defense against bacterial infection.Dev Comp Immunol. 2018 Jul;84:230-240. doi: 10.1016/j.dci.2018.02.015. Epub 2018 Feb 24. Dev Comp Immunol. 2018. PMID: 29481905
Cited by
-
Updating Our Knowledge on Fish Immunology.Int J Mol Sci. 2023 Jun 29;24(13):10856. doi: 10.3390/ijms241310856. Int J Mol Sci. 2023. PMID: 37446032 Free PMC article.
-
Bone-derived factors mediate crosstalk between skeletal and extra-skeletal organs.Bone Res. 2025 Apr 30;13(1):49. doi: 10.1038/s41413-025-00424-1. Bone Res. 2025. PMID: 40307216 Free PMC article. Review.
-
Changes in RANKL, OPG, and 25(OH)D Levels in Children with Leukemia from Diagnosis to Remission.Cancers (Basel). 2024 Aug 10;16(16):2811. doi: 10.3390/cancers16162811. Cancers (Basel). 2024. PMID: 39199584 Free PMC article.
-
Osteoclast Activation and Inflammatory Bone Diseases: Focusing on Receptors in Osteoclasts.J Inflamm Res. 2025 Mar 4;18:3201-3213. doi: 10.2147/JIR.S507269. eCollection 2025. J Inflamm Res. 2025. PMID: 40059949 Free PMC article. Review.
References
-
- Veljanovski V., Vanderbeld B., Knowles V.L., Snedden W.A., Plaxton W.C. Biochemical and Molecular Characterization of AtPAP26, a Vacuolar Purple Acid Phosphatase up-Regulated in Phosphate-Deprived Arabidopsis Suspension Cells and Seedlings. Plant Physiol. 2006;142:1282–1293. doi: 10.1104/pp.106.087171. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

