Ursodeoxycholic Acid Binds PERK and Ameliorates Neurite Atrophy in a Cellular Model of GM2 Gangliosidosis
- PMID: 37108372
- PMCID: PMC10138647
- DOI: 10.3390/ijms24087209
Ursodeoxycholic Acid Binds PERK and Ameliorates Neurite Atrophy in a Cellular Model of GM2 Gangliosidosis
Abstract
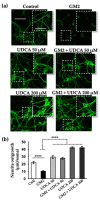
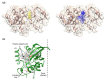
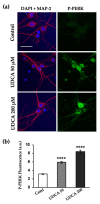
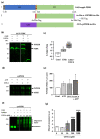
The Unfolded protein response (UPR), triggered by stress in the endoplasmic reticulum (ER), is a key driver of neurodegenerative diseases. GM2 gangliosidosis, which includes Tay-Sachs and Sandhoff disease, is caused by an accumulation of GM2, mainly in the brain, that leads to progressive neurodegeneration. Previously, we demonstrated in a cellular model of GM2 gangliosidosis that PERK, a UPR sensor, contributes to neuronal death. There is currently no approved treatment for these disorders. Chemical chaperones, such as ursodeoxycholic acid (UDCA), have been found to alleviate ER stress in cell and animal models. UDCA's ability to move across the blood-brain barrier makes it interesting as a therapeutic tool. Here, we found that UDCA significantly diminished the neurite atrophy induced by GM2 accumulation in primary neuron cultures. It also decreased the up-regulation of pro-apoptotic CHOP, a downstream PERK-signaling component. To explore its potential mechanisms of action, in vitro kinase assays and crosslinking experiments were performed with different variants of recombinant protein PERK, either in solution or in reconstituted liposomes. The results suggest a direct interaction between UDCA and the cytosolic domain of PERK, which promotes kinase phosphorylation and dimerization.
Keywords: ATP binding pocket; chemical chaperones; lysosomal storage disease.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the result.
Figures
Similar articles
-
Neurite atrophy and apoptosis mediated by PERK signaling after accumulation of GM2-ganglioside.Biochim Biophys Acta Mol Cell Res. 2019 Feb;1866(2):225-239. doi: 10.1016/j.bbamcr.2018.10.014. Epub 2018 Oct 30. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 30389374
-
[Molecular pathogenesis and therapeutic approach of GM2 gangliosidosis].Yakugaku Zasshi. 2013;133(2):269-74. doi: 10.1248/yakushi.12-00199. Yakugaku Zasshi. 2013. PMID: 23370522 Review. Japanese.
-
L-Arginine Ameliorates Defective Autophagy in GM2 Gangliosidoses by mTOR Modulation.Cells. 2021 Nov 11;10(11):3122. doi: 10.3390/cells10113122. Cells. 2021. PMID: 34831346 Free PMC article.
-
4-Phenylbutyric acid mitigates ER stress-induced neurodegeneration in the spinal cords of a GM2 gangliosidosis mouse model.Hum Mol Genet. 2025 Jan 23;34(1):32-46. doi: 10.1093/hmg/ddae153. Hum Mol Genet. 2025. PMID: 39530163 Free PMC article.
-
Animal models of GM2 gangliosidosis: utility and limitations.Appl Clin Genet. 2016 Jul 20;9:111-20. doi: 10.2147/TACG.S85354. eCollection 2016. Appl Clin Genet. 2016. PMID: 27499644 Free PMC article. Review.
Cited by
-
Effects of cholestasis and hyperammonemia on dendritic spine density and turnover in rat hippocampal neurons.Sci Rep. 2024 Dec 1;14(1):29841. doi: 10.1038/s41598-024-80871-8. Sci Rep. 2024. PMID: 39617839 Free PMC article.
-
Drug Repurposing and Lysosomal Storage Disorders: A Trick to Treat.Genes (Basel). 2024 Feb 25;15(3):290. doi: 10.3390/genes15030290. Genes (Basel). 2024. PMID: 38540351 Free PMC article. Review.
-
Potential Role of Endoplasmic Reticulum Stress in Modulating Protein Homeostasis in Oligodendrocytes to Improve White Matter Injury in Preterm Infants.Mol Neurobiol. 2024 Aug;61(8):5295-5307. doi: 10.1007/s12035-023-03905-8. Epub 2024 Jan 5. Mol Neurobiol. 2024. PMID: 38180617 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

