Genome-Wide Identification and Characterization of the Soybean Snf2 Gene Family and Expression Response to Rhizobia
- PMID: 37108411
- PMCID: PMC10138738
- DOI: 10.3390/ijms24087250
Genome-Wide Identification and Characterization of the Soybean Snf2 Gene Family and Expression Response to Rhizobia
Abstract
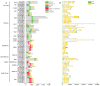
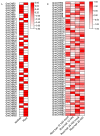
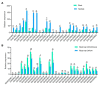
Sucrose nonfermenting 2 (Snf2) family proteins are the core component of chromatin remodeling complexes that can alter chromatin structure and nucleosome position by utilizing the energy of ATP, playing a vital role in transcription regulation, DNA replication, and DNA damage repair. Snf2 family proteins have been characterized in various species including plants, and they have been found to regulate development and stress responses in Arabidopsis. Soybean (Glycine max) is an important food and economic crop worldwide, unlike other non-leguminous crops, soybeans can form a symbiotic relationship with rhizobia for biological nitrogen fixation. However, little is known about Snf2 family proteins in soybean. In this study, we identified 66 Snf2 family genes in soybean that could be classified into six groups like Arabidopsis, unevenly distributed on 20 soybean chromosomes. Phylogenetic analysis with Arabidopsis revealed that these 66 Snf2 family genes could be divided into 18 subfamilies. Collinear analysis showed that segmental duplication was the main mechanism for expansion of Snf2 genes rather than tandem repeats. Further evolutionary analysis indicated that the duplicated gene pairs had undergone purifying selection. All Snf2 proteins contained seven domains, and each Snf2 protein had at least one SNF2_N domain and one Helicase_C domain. Promoter analysis revealed that most Snf2 genes had cis-elements associated with jasmonic acid, abscisic acid, and nodule specificity in their promoter regions. Microarray data and real-time quantitative PCR (qPCR) analysis revealed that the expression profiles of most Snf2 family genes were detected in both root and nodule tissues, and some of them were found to be significantly downregulated after rhizobial infection. In this study, we conducted a comprehensive analysis of the soybean Snf2 family genes and demonstrated their responsiveness to Rhizobia infection. This provides insight into the potential roles of Snf2 family genes in soybean symbiotic nodulation.
Keywords: ATP-dependent chromatin remodeling; Glycine max; Snf2 gene family; nodulation; rhizobia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

