The FomYjeF Protein Influences the Sporulation and Virulence of Fusarium oxysporum f. sp. momordicae
- PMID: 37108422
- PMCID: PMC10138616
- DOI: 10.3390/ijms24087260
The FomYjeF Protein Influences the Sporulation and Virulence of Fusarium oxysporum f. sp. momordicae
Abstract
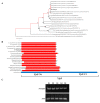
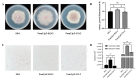
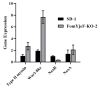
Fusarium oxysporum causes vascular wilt in more than 100 plant species, resulting in massive economic losses. A deep understanding of the mechanisms of pathogenicity and symptom induction by this fungus is necessary to control crop wilt. The YjeF protein has been proven to function in cellular metabolism damage-repair in Escherichia coli and to play an important role in Edc3 (enhancer of the mRNA decapping 3) function in Candida albicans, but no studies have been reported on related functions in plant pathogenic fungi. In this work, we report how the FomYjeF gene in F. oxysporum f. sp. momordicae contributes to conidia production and virulence. The deletion of the FomYjeF gene displayed a highly improved capacity for macroconidia production, and it was shown to be involved in carbendazim's associated stress pathway. Meanwhile, this gene caused a significant increase in virulence in bitter gourd plants with a higher disease severity index and enhanced the accumulation of glutathione peroxidase and the ability to degrade hydrogen peroxide in F. oxysporum. These findings reveal that FomYjeF affects virulence by influencing the amount of spore formation and the ROS (reactive oxygen species) pathway of F. oxysporum f. sp. momordicae. Taken together, our study shows that the FomYjeF gene affects sporulation, mycelial growth, pathogenicity, and ROS accumulation in F. oxysporum. The results of this study provide a novel insight into the function of FomYjeF participation in the pathogenicity of F. oxysporum f. sp. momordicae.
Keywords: Fusarium oxysporum; YjeF protein; sporulation; virulence.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Sun S.K., Huang J.W. A new Fusarium wilt of bitter gourd in Taiwan. Plant Dis. 1983;67:226–227. doi: 10.1094/PD-67-226. - DOI
-
- Shih J., Wei Y., Goodwin P.H. A comparison of the pectate lyase genes, pel-1 and pel-2, of Colletotrichum gloeosporioides f. sp. malvae and the relationship between their expression in culture and during necrotrophic infection. Gene. 2000;243:139–150. doi: 10.1016/S0378-1119(99)00546-6. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

