Automatic Generation of SBML Kinetic Models from Natural Language Texts Using GPT
- PMID: 37108453
- PMCID: PMC10138937
- DOI: 10.3390/ijms24087296
Automatic Generation of SBML Kinetic Models from Natural Language Texts Using GPT
Abstract
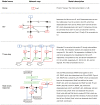
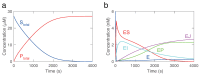
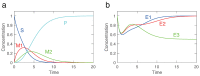
Kinetic modeling is an essential tool in systems biology research, enabling the quantitative analysis of biological systems and predicting their behavior. However, the development of kinetic models is a complex and time-consuming process. In this article, we propose a novel approach called KinModGPT, which generates kinetic models directly from natural language text. KinModGPT employs GPT as a natural language interpreter and Tellurium as an SBML generator. We demonstrate the effectiveness of KinModGPT in creating SBML kinetic models from complex natural language descriptions of biochemical reactions. KinModGPT successfully generates valid SBML models from a range of natural language model descriptions of metabolic pathways, protein-protein interaction networks, and heat shock response. This article demonstrates the potential of KinModGPT in kinetic modeling automation.
Keywords: GPT; kinetic modeling; large language model; simulation; systems biology.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures











Similar articles
-
SBML-PET: a Systems Biology Markup Language-based parameter estimation tool.Bioinformatics. 2006 Nov 1;22(21):2704-5. doi: 10.1093/bioinformatics/btl443. Epub 2006 Aug 22. Bioinformatics. 2006. PMID: 16926221
-
CSML2SBML: a novel tool for converting quantitative biological pathway models from CSML into SBML.Biosystems. 2014 Jul;121:22-8. doi: 10.1016/j.biosystems.2014.05.004. Epub 2014 Jun 2. Biosystems. 2014. PMID: 24881961
-
Evolving a lingua franca and associated software infrastructure for computational systems biology: the Systems Biology Markup Language (SBML) project.Syst Biol (Stevenage). 2004 Jun;1(1):41-53. doi: 10.1049/sb:20045008. Syst Biol (Stevenage). 2004. PMID: 17052114 Review.
-
The Systems Biology Markup Language (SBML): Language Specification for Level 3 Version 2 Core Release 2.J Integr Bioinform. 2019 Jun 20;16(2):20190021. doi: 10.1515/jib-2019-0021. J Integr Bioinform. 2019. PMID: 31219795 Free PMC article.
-
Modeling and Simulation Tools: From Systems Biology to Systems Medicine.Methods Mol Biol. 2016;1386:441-63. doi: 10.1007/978-1-4939-3283-2_19. Methods Mol Biol. 2016. PMID: 26677194 Review.
References
-
- Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., Arkin A.P., Bornstein B.J., Bray D., Cornish-Bowden A., et al. The systems biology markup language (SBML): A medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

