Identification of Signature Genes of Dilated Cardiomyopathy Using Integrated Bioinformatics Analysis
- PMID: 37108502
- PMCID: PMC10139023
- DOI: 10.3390/ijms24087339
Identification of Signature Genes of Dilated Cardiomyopathy Using Integrated Bioinformatics Analysis
Abstract
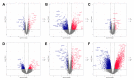
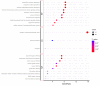
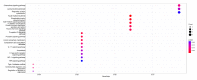
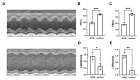
Dilated cardiomyopathy (DCM) is characterized by left ventricular or biventricular enlargement with systolic dysfunction. To date, the underlying molecular mechanisms of dilated cardiomyopathy pathogenesis have not been fully elucidated, although some insights have been presented. In this study, we combined public database resources and a doxorubicin-induced DCM mouse model to explore the significant genes of DCM in full depth. We first retrieved six DCM-related microarray datasets from the GEO database using several keywords. Then we used the "LIMMA" (linear model for microarray data) R package to filter each microarray for differentially expressed genes (DEGs). Robust rank aggregation (RRA), an extremely robust rank aggregation method based on sequential statistics, was then used to integrate the results of the six microarray datasets to filter out the reliable differential genes. To further improve the reliability of our results, we established a doxorubicin-induced DCM model in C57BL/6N mice, using the "DESeq2" software package to identify DEGs in the sequencing data. We cross-validated the results of RRA analysis with those of animal experiments by taking intersections and identified three key differential genes (including BEX1, RGCC and VSIG4) associated with DCM as well as many important biological processes (extracellular matrix organisation, extracellular structural organisation, sulphur compound binding, and extracellular matrix structural components) and a signalling pathway (HIF-1 signalling pathway). In addition, we confirmed the significant effect of these three genes in DCM using binary logistic regression analysis. These findings will help us to better understand the pathogenesis of DCM and may be key targets for future clinical management.
Keywords: DEGs; RNA-Seq; dilated cardiomyopathy; microarray; robust rank aggregation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

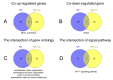
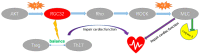
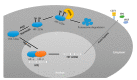
Similar articles
-
Integrated microarray analysis to identify potential biomarkers and therapeutic targets in dilated cardiomyopathy.Mol Med Rep. 2020 Aug;22(2):915-925. doi: 10.3892/mmr.2020.11145. Epub 2020 May 14. Mol Med Rep. 2020. PMID: 32626989 Free PMC article.
-
An Robust Rank Aggregation and Least Absolute Shrinkage and Selection Operator Analysis of Novel Gene Signatures in Dilated Cardiomyopathy.Front Cardiovasc Med. 2021 Dec 14;8:747803. doi: 10.3389/fcvm.2021.747803. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 34970603 Free PMC article.
-
Nuclear damage-induced DNA damage response coupled with IFI16-driven ECM remodeling underlies dilated cardiomyopathy.Theranostics. 2025 Apr 28;15(12):5998-6021. doi: 10.7150/thno.112247. eCollection 2025. Theranostics. 2025. PMID: 40365289 Free PMC article.
-
Characteristic adaptations of the extracellular matrix in dilated cardiomyopathy.Int J Cardiol. 2016 Oct 1;220:634-46. doi: 10.1016/j.ijcard.2016.06.253. Epub 2016 Jun 27. Int J Cardiol. 2016. PMID: 27391006 Review.
-
Inflammatory dilated cardiomyopathy (DCMI).Herz. 2005 Sep;30(6):535-44. doi: 10.1007/s00059-005-2730-5. Herz. 2005. PMID: 16170686 Review.
Cited by
-
Novel molecular mechanisms of immune evasion in hepatocellular carcinoma: NSUN2-mediated increase of SOAT2 RNA methylation.Cancer Commun (Lond). 2025 Jul;45(7):846-879. doi: 10.1002/cac2.70023. Epub 2025 Apr 14. Cancer Commun (Lond). 2025. PMID: 40227950 Free PMC article.
-
Unraveling pathogenesis, biomarkers and potential therapeutic agents for endometriosis associated with disulfidptosis based on bioinformatics analysis, machine learning and experiment validation.J Biol Eng. 2024 Jul 26;18(1):42. doi: 10.1186/s13036-024-00437-0. J Biol Eng. 2024. PMID: 39061076 Free PMC article.
-
Integrative Approaches to Myopathies and Muscular Dystrophies: Molecular Mechanisms, Diagnostics, and Future Therapies.Int J Mol Sci. 2025 Aug 18;26(16):7972. doi: 10.3390/ijms26167972. Int J Mol Sci. 2025. PMID: 40869293 Free PMC article. Review.
-
Bioinformatic identification of important roles of COL1A1 and TNFRSF12A in cartilage injury and osteoporosis.J Int Soc Sports Nutr. 2025 Dec;22(1):2454641. doi: 10.1080/15502783.2025.2454641. Epub 2025 Jan 23. J Int Soc Sports Nutr. 2025. PMID: 39847474 Free PMC article.
References
-
- Richardson P., McKenna W., Bristow M., Maisch B., Mautner B., O’Connell J., Olsen E., Thiene G., Goodwin J., Gyarfas I., et al. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation. 1996;93:841–842. doi: 10.1161/01.cir.93.5.841. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

