HP1γ Prevents Activation of the cGAS/STING Pathway by Preserving Nuclear Envelope and Genomic Integrity in Colon Adenocarcinoma Cells
- PMID: 37108510
- PMCID: PMC10138453
- DOI: 10.3390/ijms24087347
HP1γ Prevents Activation of the cGAS/STING Pathway by Preserving Nuclear Envelope and Genomic Integrity in Colon Adenocarcinoma Cells
Abstract
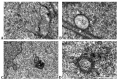
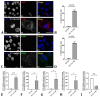
Chronic inflammatory processes in the intestine result in serious conditions such as inflammatory bowel disease (IBD) and cancer. An increased detection of cytoplasmic DNA sensors has been reported in the IBD colon mucosa, suggesting their contribution in mucosal inflammation. Yet, the mechanisms altering DNA homeostasis and triggering the activation of DNA sensors remain poorly understood. In this study, we show that the epigenetic regulator HP1γ plays a role in preserving nuclear envelope and genomic integrity in enterocytic cells, thereby protecting against the presence of cytoplasmic DNA. Accordingly, HP1 loss of function led to the increased detection of cGAS/STING, a cytoplasmic DNA sensor that triggers inflammation. Thus, in addition to its role as a transcriptional silencer, HP1γ may also exert anti-inflammatory properties by preventing the activation of the endogenous cytoplasmic DNA response in the gut epithelium.
Keywords: HP1γ; STING; cGAS; cytosolic DNA; epigenetics; inflammation; inflammatory bowel disease; nuclear envelop.
Conflict of interest statement
The authors declare that there is no competing interest associated with the manuscript. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




Similar articles
-
cGAS-STING DNA-sensing in inflammatory bowel diseases.Trends Mol Med. 2025 Feb;31(2):165-180. doi: 10.1016/j.molmed.2024.10.002. Epub 2024 Oct 23. Trends Mol Med. 2025. PMID: 39448330 Review.
-
Mitochondrial DNA drives noncanonical inflammation activation via cGAS-STING signaling pathway in retinal microvascular endothelial cells.Cell Commun Signal. 2020 Oct 28;18(1):172. doi: 10.1186/s12964-020-00637-3. Cell Commun Signal. 2020. PMID: 33115500 Free PMC article.
-
Mitochondrial DNA leakage induces odontoblast inflammation via the cGAS-STING pathway.Cell Commun Signal. 2021 May 20;19(1):58. doi: 10.1186/s12964-021-00738-7. Cell Commun Signal. 2021. PMID: 34016129 Free PMC article.
-
DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway.Proc Natl Acad Sci U S A. 2017 Nov 14;114(46):12196-12201. doi: 10.1073/pnas.1708744114. Epub 2017 Oct 30. Proc Natl Acad Sci U S A. 2017. PMID: 29087318 Free PMC article.
-
cGAS-STING signaling pathway in gastrointestinal inflammatory disease and cancers.FASEB J. 2022 Jan;36(1):e22029. doi: 10.1096/fj.202101199R. FASEB J. 2022. PMID: 34907606 Review.
Cited by
-
cGAS/STING pathway and gastrointestinal cancer: Mechanisms and diagnostic and therapeutic targets (Review).Oncol Rep. 2025 Jan;53(1):15. doi: 10.3892/or.2024.8848. Epub 2024 Nov 29. Oncol Rep. 2025. PMID: 39611480 Free PMC article. Review.
-
How Does cGAS Avoid Sensing Self-DNA under Normal Physiological Conditions?Int J Mol Sci. 2023 Sep 29;24(19):14738. doi: 10.3390/ijms241914738. Int J Mol Sci. 2023. PMID: 37834184 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

