Network Biology Analyses and Dynamic Modeling of Gene Regulatory Networks under Drought Stress Reveal Major Transcriptional Regulators in Arabidopsis
- PMID: 37108512
- PMCID: PMC10139068
- DOI: 10.3390/ijms24087349
Network Biology Analyses and Dynamic Modeling of Gene Regulatory Networks under Drought Stress Reveal Major Transcriptional Regulators in Arabidopsis
Abstract
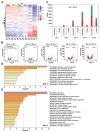
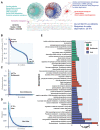
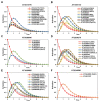
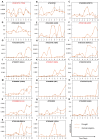
Drought is one of the most serious abiotic stressors in the environment, restricting agricultural production by reducing plant growth, development, and productivity. To investigate such a complex and multifaceted stressor and its effects on plants, a systems biology-based approach is necessitated, entailing the generation of co-expression networks, identification of high-priority transcription factors (TFs), dynamic mathematical modeling, and computational simulations. Here, we studied a high-resolution drought transcriptome of Arabidopsis. We identified distinct temporal transcriptional signatures and demonstrated the involvement of specific biological pathways. Generation of a large-scale co-expression network followed by network centrality analyses identified 117 TFs that possess critical properties of hubs, bottlenecks, and high clustering coefficient nodes. Dynamic transcriptional regulatory modeling of integrated TF targets and transcriptome datasets uncovered major transcriptional events during the course of drought stress. Mathematical transcriptional simulations allowed us to ascertain the activation status of major TFs, as well as the transcriptional intensity and amplitude of their target genes. Finally, we validated our predictions by providing experimental evidence of gene expression under drought stress for a set of four TFs and their major target genes using qRT-PCR. Taken together, we provided a systems-level perspective on the dynamic transcriptional regulation during drought stress in Arabidopsis and uncovered numerous novel TFs that could potentially be used in future genetic crop engineering programs.
Keywords: Arabidopsis; co-expression network; computational simulation; drought; network centrality; systems biology; transcriptional regulation; water deprivation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Abiotic stress induced miRNA-TF-gene regulatory network: A structural perspective.Genomics. 2020 Jan;112(1):412-422. doi: 10.1016/j.ygeno.2019.03.004. Epub 2019 Mar 12. Genomics. 2020. PMID: 30876925
-
Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants.Proc Natl Acad Sci U S A. 2018 Jun 19;115(25):6494-6499. doi: 10.1073/pnas.1721487115. Epub 2018 May 16. Proc Natl Acad Sci U S A. 2018. PMID: 29769331 Free PMC article.
-
A transcriptional dynamic network during Arabidopsis thaliana pollen development.BMC Syst Biol. 2011;5 Suppl 3(Suppl 3):S8. doi: 10.1186/1752-0509-5-S3-S8. Epub 2011 Dec 23. BMC Syst Biol. 2011. PMID: 22784627 Free PMC article.
-
Insights into molecular links and transcription networks integrating drought stress and nitrogen signaling.New Phytol. 2024 Jan;241(2):560-566. doi: 10.1111/nph.19403. Epub 2023 Nov 16. New Phytol. 2024. PMID: 37974513 Review.
-
Transcriptional gene network involved in drought stress response: application for crop breeding in the context of climate change.Philos Trans R Soc Lond B Biol Sci. 2025 May 29;380(1927):20240236. doi: 10.1098/rstb.2024.0236. Epub 2025 May 29. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 40439309 Free PMC article. Review.
Cited by
-
Time-course transcriptome analysis reveals gene co-expression networks and transposable element responses to cold stress in cotton.BMC Genomics. 2025 Mar 12;26(1):235. doi: 10.1186/s12864-025-11433-z. BMC Genomics. 2025. PMID: 40075303 Free PMC article.
-
Microbiome Structures and Beneficial Bacteria in Soybean Roots Under Field Conditions of Prolonged High Temperatures and Drought Stress.Microorganisms. 2024 Dec 19;12(12):2630. doi: 10.3390/microorganisms12122630. Microorganisms. 2024. PMID: 39770832 Free PMC article.
-
Integrated Systems Biology Pipeline to Compare Co-Expression Networks in Plants and Elucidate Differential Regulators.Plants (Basel). 2023 Oct 19;12(20):3618. doi: 10.3390/plants12203618. Plants (Basel). 2023. PMID: 37896081 Free PMC article.
-
Plasma Optimization as a Novel Tool to Explore Plant-Microbe Interactions in Climate Smart Agriculture.Microorganisms. 2025 Jan 13;13(1):146. doi: 10.3390/microorganisms13010146. Microorganisms. 2025. PMID: 39858915 Free PMC article. Review.
-
Profiling of Key Hub Genes Using a Two-State Weighted Gene Co-Expression Network of 'Jao Khao' Rice under Soil Salinity Stress Based on Time-Series Transcriptome Data.Int J Mol Sci. 2024 Oct 16;25(20):11086. doi: 10.3390/ijms252011086. Int J Mol Sci. 2024. PMID: 39456877 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

